"Helix-Forming Carbohydrate Amino Acids", by T. D. W. claridge, D. D. Long, C. M. Baker, B. Odell, G. H. Grant, A. A. Edwards, G. E. Tranter, G. W. J. Fleet, M. D. Smith, Journal of Organic Chemistry, March 18 2005, 70, 6, 2082 - 2090
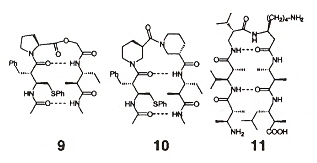
Protein-like secondary folding can be found in synthetic peptide-like macromolecules based upon Carbohydrate Amino Acids. β-peptides have already been shown to have helical, turn, and sheet conformations. For example, β-peptides based upon oxetane (ring ether) adopt 10-helical comnformations, thus establishing one area of peptidomimetics of interest.
|
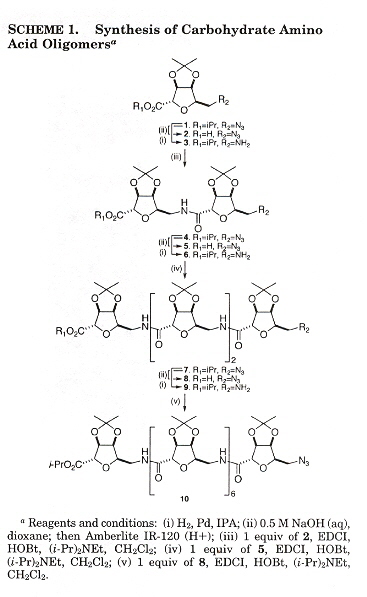
Carbohydrate Amino Acid Oligomers
|
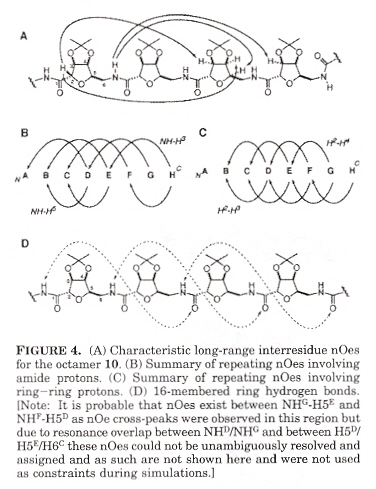
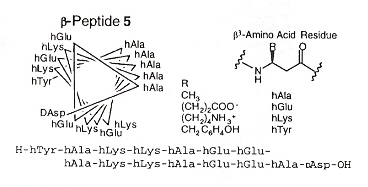
Helix Structure of Carbohydrate Amino Acids
|
"β-Peptides: From Structure to Function", by R. P Cheng, S. H. Gellman, W. F. DeGrado, Chemical Reviews, 2001, 101, 3219 - 3232
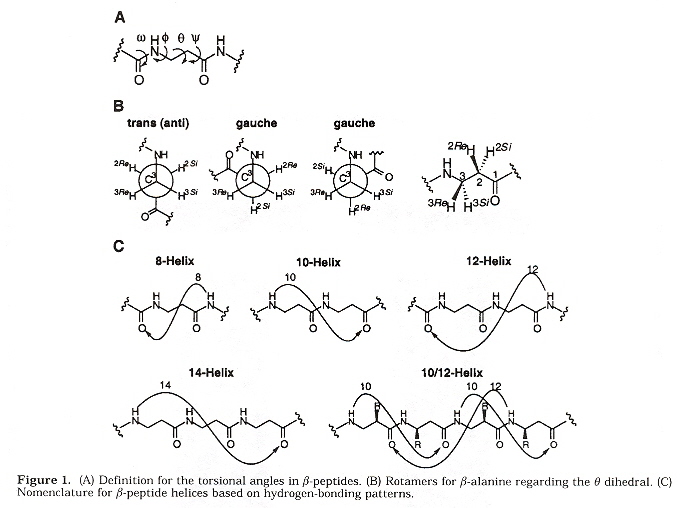
This paper is an overview dealing with the possibilities of designing biomimetic polymers with both secondary and tertiary structures. A few words about peptides: α-peptides are composed of α-amino acids. Similarly, β-peptides are composed of β-amino acids.
The conformations of β-peptides depend upon the main chain torsional angles ω, φ, θ, and ψ (Balaram convention). β-peptide sheet secondary structures come in two forms: anti C2-C3 torsion angle, and "gauche" C2-C3 torsion angle. The "anti" β-peptide sheet has a net dipole due to carbonyls oriented in the same direction. The "gauche" type has no net dipole.
|
Effects of Torsion angle θ
|
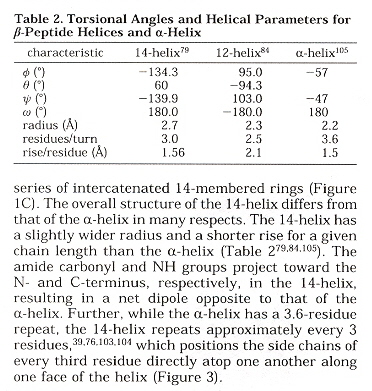
Torsion Angles and Helices
|
|
Down the throat of the Helix
|
β-sheets
|
"High solubility of random-sequence proteins consisting of five kinds of primitive amino acids", N. Doi, K. Kakukawa, Y. Oishi, H. Yanagawa, Protein Engineering, Design & Selection, May 31, 2005, 18, 6, 279-284
Early proteins evolved from a reduced alphabet of the 20 standard amino acids. Proteins composed only of Ala, Gly, Val, Asp, and Glu (believed to have been the most abundant in the prebiotic environment) have a higher solubility given the same level of hyrophobicity as proteins incorporating the 20-alphabet amino acids.
"A gamut of loops: meandering DNA", by S. Semsey, K. Varnik, S. Adhya, TRENDS in Biochemical Sciences, June 2005, 30, 6, 334 - 340
An interesting discussion concerning how DNA loops, under the geometric effects of proteins. This discussion proceeds along lines discussed in "Emergent Computation: Emphasizing Bioinformatics" in Chapter 5, pages 166 - 183.
"A monoclonal antibody that inhibits mycobacterial DNA gyrase by a novel mechanism", by U. H. Manjunatha, A. Maxwell, V. Nagaraja, Nucleic Acids Research, June 1, 2005, 33, 10, 3085 - 3084
A new mechanism that inhibits DNA gyrase (a type II supercoiling topoisomerase) activity in Mycobacterium smegmatis and Mycobacterium tuberculosis is described. Two GyrA and GyrB units bind duplex DNA where one segment named the transported "T" segment lies over the other gated or "G" segment. The gyrase transiently cleaves the G segment and transports the T segment through this break, then religation occurs. Specific conformational steps of topological significance are described. An interesting discussion concerning how DNA loops, under the geometric effects of proteins. This discussion proceeds along lines discussed in "Emergent Computation: Emphasizing Bioinformatics" in Chapter 5, pages 166 - 183.
"Single-Molecule RNA Folding", by G. Bokinsky, X. Zhuang, Accounts of Chemical Research, July, 2005, 38, 7, 566 - 573
The conformational dynamics of RNA secondary folding using FRET to study small single-molecules of RNA. Specifically, a hairpin ribozyme (RNA enzyme) is studied and the detailed mechanisms inferred. Cy3 fluorescent donor at the ribozyme and Cy5 fluorescent acceptor are used to study the folding action and the final effect on a substrate "S". The structural studies are used to infer pathways composed of docking (folding), cleavage, and undocking (unfolding) result in cleaved products. Different rate constants correspond to possible different folding conformations. The large multidomain ribozyme for Tetrahymena is also studied to elucidate at least three different dynamic folding pathways.
"Orientation Control of Fluorescence Resonance Energy Transfer Using DNA as a Helical Scaffold", by F. D. Lewis, L. Zhang, X. Zuo, Journal of the American Chemical Society, July 20, 2005, 127, 10002 - 10003
Fluorescence Resonance Energy Transfer (FRET) consists of attaching fluroescent donor and fluorescent acceptor dyes at two specific sites of a biomolecule. Energy transfer takes place between each dye resulting in a decrease in donor emission accompanied by an increase in acceptor emission. Emission varies as a consequence of changes in molecular conformation such as the distance "R" between the donor and the acceptor using the Förster radius "R0". Specifically, "SA" (stilbene dicarboxamide) is used as a donor, and "PA" (perylenedicarboxamide) is used as a donor. Spectra of the fluorescence emissions of this acceptor/donor pair are studied in different A-T stackings.
"Docking kinetics and equilibrium of a GAAA tetraloop-receptor motif probed by single-molecule FRET", by J. H. Hodak, C. D. Downey, J. L. Fiore, A. Pardi, D. J. Nesbitt, Proceedings of the National Academy of Sciences of the USA", July 26 2005, 102, 30, 10505 - 10510
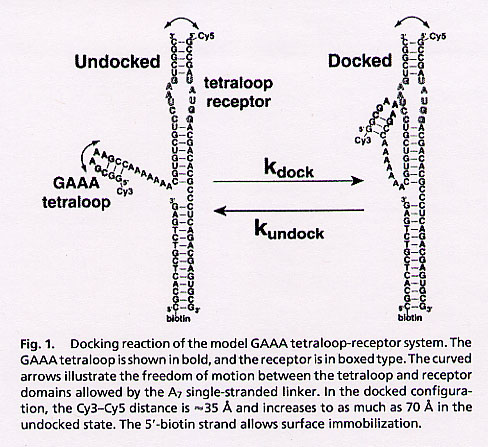
An RNA system is designed that contains a single GAAA tetraloop and a single tetraloop receptor. The tetraloop and its receptor are joined by a flexible single-stranded linker that isolates tertiary folding from other tertiary effects. FRET dyes differentiate unfolded from tertiary folded states, thereby supporting kinetic and equilibrium studies.