"DNA methylation and cancer therapy: new developments and expectations", by M. Esteller, Current Opinion in Oncology, January, 2005, 17, 5, 55 - 60
Epigenetics (inheritance based upon gene-expression independent of the underlying DNA nucleotide sequence), as opposed to genetics (inheritance based upon the DNA nucleotide sequence) is studied. Methylated DNA is inherited at the cellular level during cell division, thus the term "epigenetics". The main form of epigenetics studied in this paper is methylation of cytosine (C) when preceding guanine (G) forming the dinucleotide CpG "island". The specific issue of interest is the mechanisms of DNA-demethylating agents used in anticancer drugs.
"The (Dual) Origin of Epigenetics", by D. Haig, Cold Spring Harbor Symposia on Quantitative Biology, 2004, 69, 67 - 70
The word Epigenetics was first used by Waddingtona to refer to the study of "causal mechanisms" by which "the genes of the genotype bring about phenotypic effects"b. Waddington says "...by the time development begins, the zygote contains certain 'preformed' characters, but that these must interact with one another, in process of 'epigenesis', before the adult condition is attained. The study of the 'preformed' characters nowadays belongs to the discipline known as 'genetics'; the name 'epigenetics' is suggested as the study of those processes which constitute the epigenesis which is also involved in development"c. Thus Waddington's "epigenetics" means "developmental biology". Waddington felt that many geneticists held a naive view that genes corresponded to characters. Instead, development for Waddington was determined by the interaction of many genes along with the environment. Waddington points out that "genetics" often ignores the process of development as it studies life in its simplest form (before much of the processes of development take place). Waddington underlines this when he refers to Haecker's use of the term "phenogenetics". Neo-Darwinism in Waddington's view, involved a "breach between organism and nature as complete as the Cartesian dualism of mind and matter..."d.
"Epigenetics" was used by Nanneye in the belief that "auxilliary mechanisms with different principles of operation are involved in determining which specificities are to be expressed in any particular cell." Specifically, Nanney felt that cells with the same genotype could have different phenotypes. Nanney felt that there were two opposing viewpoints: those who ascribed a predominant role to heredity, evolution, and genetic material of the nucleus (DNA) as opposed to those who felt that extranuclear, cytoplasmic factors were also significantf. Nanney was also concerned about the interaction of science and political philosophy. The concept of the "master molecule" in the chromosomes is the basis of the organism's characteristics, all other cellular constituents are considered relatively inconsequential: a view much like that of a totalitarian government (see the viewpoint of Bussard in another paper dealing with prions and retroviruses). Nanney felt that the cytoplasm also contained molecules important to inheritance, in equillibrium at "steady state"g.
a "The epigenotype", by C. H. Waddington, Endeavour, 1942, 1, 18
b An introduction to modern genetics, byC. H. Waddington, Macmillan, 1939, New York, p. 156
c ibid, pp. 154, 155
d The strategy of the genes, by C. H. Waddington, George Allen & Unwin, 1953, London, p. ix
e "Epigenetic control systems", Proceedings of the National Academy of Sciences U. S., 1958, 44, 7, 712
f The chemical basis of heredity, edited by W. D. McElroy and B. Glass, Johns Hopkins University Press, Baltimore, Maryland, 1957, pp. 134, 136
g "Epigenetic factors affecting mating type expression in certain ciliates", Cold Spring Harbor Symposia on Quantitative Biology, 1959, 23, 327
"Epigenetic Mechanisms in Early Mammalian Development", by D. Solter, T. Hiiragi, A. V. Evsikov, J. Moyer, W. N. De Vries, A. E. Peaston, B. B. Knowles, Cold Spring Harbor Symposia on Quantitative Biology, 2004, 69, 11 - 17
The informational content of the mammalian egg at fertilization is not restricted to its DNA sequence, but also to DNA and chromatin modifications (histones and protamines), other macromolecules such as RNAs and proteins, and possibly the cytoplasm and plasma membrane. Upon fertilization, the paternal genome is rapidly demethylated, while the maternal genome is gradually demethylated. Differences between paternally and maternally derived chromatin persist until the two-cell blastomere (lost by the next cellular division: the four-cell blastomere). As complex patterns of imprinted genes by metylation of DNA is limited due to stability considerations, the histones that wrap around the DNA support the capability of far more complex patterns of marking (and do not change their locus). Histone and chromatin marking include methylation, phosphorylation, and acetylationa, b. mRNA concentrations fluctuate during early embryonic development (when there is supposedly no transcription). Reverse transcriptase activity is essential during embryonic development until the four-cell stage due to reverse transcriptase inhibitor nevoparine. Transposable elements can create mutations which can activate and silence genes. At fertilization, a unique situation exists where one cell exists but with two nuclei, thus there is a possibility of the first cleavage plane supporting a lack of bilateral symmetry. It has been assumed that at the two-cell bastomere stage, one cell containes the sperm entry point and thus can develop differently from the other cell. All these structures and molecules permit differential epigenetic development.
a "Epigenetic Regulation by Histone Methylation and Histone Variants", by P. Cheung, P. Lau, Molecular Endocrinology, March 2005, 19, 3, 563 - 573
b "Chromatin remodelling and Epigenetic features of germ cells", by S. Kimmins, P. Sassone-Corsi, Nature, March 31 2005, 434, 7033, 583 - 589
"DNA Methylation and Human Disease", by K. D. Robertson, Nature Reviews Genetics, Aug. 2005, 6, 8, 597 - 610
A very interesting review about the role of methylation in human diseases. Mammalian DNA methylation is composed of two components, DNMTs (DNA methyltransferases) concerned with establishing and maintaining DNA methylation patterns, and MBDs (methyl-CpG binding proteins) which "read" or interpret methylation marks. CpG islands are regions of approximatly 1K bases rich in G–C content, rich in CpG dinucleotides, usually hypomethylated. An interesting dual mechanism seems to underlie cancer: Hypermethylation at CpG islands causes cellular growth, while genes associated with DNA repair and apoptosis are hypermethylated (are under expressed)! Cancer cells proliferate due to DNA damage (methylation) while this damage turns off repair processes or process that would kill these cancerous cells.
Autoimmune diseases sometimes are caused by T cells which have reuced DNA methylation, thus are over expressed.
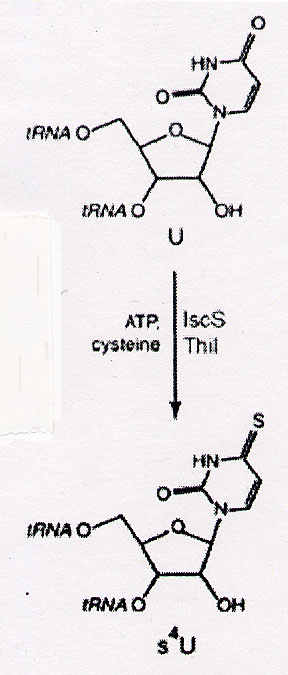
"A novel DNA modification by sulphur", by X. Zhou, X. He, J. Liang, A. Li, T. Xu, T. Kieser, J. D. Helmann, Z. Deng, Molecular Biology, Sept. 2005, 57, 5, 1428 - 1438
While methylation is the most common covalent modification of DNA, DNA has also been found to be modified at G-specific sites. Study indicated that these modifications involved the incorporation of sulphur or a sulphur containing substance into DNA. 4-thiouridine. The predominant thionucleotide in E. Coli tRNA is 4-thiouridine s4U (see below).