"The Genetic Language: Grammar, Semantics, Evolution", by V. A. Ratner, May 1993, 29, 5, 709 - 719
This very important paper dicusses a number of very importat topics in the relationship between Language and genetics. It is to be noted that the author is correctly very interested in both semantics and evolution.
The author points out that semantics includes the following:
- "Functional traits of the structure of macromolecules", thus semantics corresponds to chemical forces and steric conformations.
- The correspondence between codons and their encoded amino acids.
- Seconadry structures (of proteins and RNA). Hydrophily and hydrophoby of amino acids, etc.
- Just as there are exons and introns, semantics also extends to fibrinopeptide excision of segments of proteins.
- Consensus has a semantic nature (exact molecular composition may not be required, just a stochastic distribution, as in proteins).
Thus 'semantics' in a genetics setting radically departs from mental, pschological restrictions of human languages.
The author extends applications of liguistics to include:
- Codons
- Cistrons (two or more dependent "genes" [+, +] or [–, –] or units that together act like a gene when on the same chromosome or genomic bodies)
- Replicons
- Plasmids
- Scriptons (operons: a group of genes that act together)
- Exons and introns
- Mobile elements (genomic units that are highly transmissable and mobile or promiscuous, in organelles such as mitochondria, chloroplasts, etc. and plasmids, with different dialects)
- Linkage maps
- Endonuclease sites, methytransferases, topoisomerization enzymes, gyrases, telomeres (quadruplexes), etc.
Examples of the importance of evolution include that linguistics applies to cystrons and proteins in a prebiotic stage; the limitation to 20 amino acids occurs after physical-chemical interactions with oligonuleotides. The linguistic rules that apply to RNA secondary structure developes after Watson-Crick complementary bases have appeared. In addition to the order of evolutionary development, selection also operates. The author points out that evolutionary rate also can be examined: "the rules of genetic language (only one!) are evolving too, but much more slowly than genetic codes." Note that linguistic evolution semantically depends upon molecular evolution.
Areas where others hold opinions that differ from those expressed in this paper, include:
- The viewpoint is restricted to genetic languages, excluding non-genetic applications of biochemistry and chemistry of macromolecules (for example: nanotechnology, and artificial bases, artificial zinc finger endonucleases and methyltransferases, artificial or synthetic proteins containing amino acids not found in nature, some β–amino acids, PNA in lieu of DNA, etc.).
- Genetic texts (where language applies) are restricted to "linear ordered symbol sequences", but 2-dimensional and 3-dimensional languages (for example, shape grammars) transcend this restriction.
- Hierarchical structure typically is understood to be a tree-structure, but non-tree structured grammars are required, for example, in dealing with pseudoknots.
- Consensus might apply in other areas, such as prions, and other epigenetic aspects.
- There is only one genetic language.
Two areas discussed in this paper are extremely significant and radically depart from the views expressed by leading linguists such as N. Chomsky:
- Semantics is not limited to human languages.
- Evolution of linguistic grammars must be explicitly treated, and language theory cannot violate this requirement of nature (Chomsky excludes the evolutionary development from simple to the more complex, viewing simple languages as not counting as languages).
No actual language or grammar is explicitly presented in this paper.
"Block-Modular Principle of Organization and Evolution of Molecular-Genetic Regulatory Systems (MGRS)", by V. A. Ratner, Genetika, May 1992, 28, 2, 5 - 24
The viewpoint expressed in this paper is that "hierarchy" and "evolution" acts upon block-modules at all levels of life, including local elaboration, replacement, and repair. Ratner points out that genetic modification is based upon the block-modular organization also a, b. Examples where a modular organization are found, include the following.
- Organisms, including the genome of the organism.
- Tissues, organs, and genetic systems that pertain to tissues and organs.
- Cells, subsytems of cells, and genetic systems that pertain to cells.
- Organelles and chromosomes, and genetic systems that pertain to organelles and chromosomes.
- Aggregations of proteins, ribosomes, operons, genes, etc.
- Domains of proteins and genes, such as exons, introns, junk sequences, stretches of linker oligonucleotides and linker proteins, etc.
Ratner points out that an alternative to block-modular organization is a distributed organization (both designs of organization in fact applying). In the case of distributed organization, the number of components influences intensity of expression. Distributed organization is found in communities, populations, ecosystems, for example.
Ratner goes on to analyze specific areas that show evidence of MGRS, as in macromolecules including zinc-fingers, codons, protein domains such as Go's modules, Exons/Introns, genetic sites such as operators, Shine-Delgarno sites, noncoding regions, phage genomes, etc.
a "Evolution and Tinkering", by F. Jacob, Science, 1977, 196, 1161 - 1166
b "THE ROLE OF MOBILE GENETIC ELEMENTS (MGE) IN MICROEVOLUTION", by V. A. Ratner, L. A. Vasilyeva, Genetica, May - Dec. 1992, 28, 12, 5 - 17
"Grammatical analysis of DNA sequences provides a rationale for the regulatory control of an entire chromosome", by S. Ohno, Genetic Research, Oct.-Dec. 1990, 56, 2-3, 115 - 120
This paper is concerned with sets of sequences of DNA bases. Thus given the four bases, A, C, G, T and the Watson-Crick relationship, there are 42 = 16 possible sequences of two bases. Similarly there are 43 = 64 possible sequences of three bases. Considered are 44 = 256 sequences of 4 bases, 46 possible sequences of six bases, etc. These sequences have a semantic interpretation, thus sequences of three bases correspond to codons. Semantically, palindromic sequences of 7 to 8 bases are often targets for DNA-binding proteins. Also, from a semantic point of view, the palindromic tetramer "TATA" serves as the transcription initiation site promotors for RNA polymerase II. Sequences with semantic value are especially noted "grammatical" rules (corresponding to grammatical words in common laguages, such as the 3-base rule that corresponds to the word "the" in English). Thus the heptamer "CAGNGTG" has various semantic values in components of immunoglobulin:
- N = A, then in the 3' coding region: VL, VH, and DH
- N = T, then in the 5' coding region: JL, JH, and DG
The author considers statistical distributions of syllables of two bases. Of the 42 = 16 possible syllables of two bases, TG, CT, and CA appear with high frequency in all DNA, while CG and TA appear with very low frequency. Important relationships are singled out:
- TG has Watson-Crick complementary syllable CA
- CG corresponds to palindrome syllable CG, TA corresponds to palindrome syllabel TA
Thus for the 16 syllables composed of two bases, Human serum albumin gene and insectivore immunoglobulin IgCμH, the following distributions are considered (the middle column being mirror image reversals of the first and third columns, xy & yx), the blue dimers occurring in deficient or low distribution.
| TG | GT-- |
| CT | TC-- |
| CA | AC-- |
| -- | GCCG |
| -- | ATTA |
Junk sequences in DNA are also discussed. No actual grammar is explicitly presented in this paper.
"The grammatical rule for all DNA: Junk and coding sequences", by S. Ohno, T. Yomo, Electrophoresis, Feb.-Mar. 1991, 12, 2-3, 103 - 108
DNA and RNA in the pre-biotic world are examined. The auhtors conclude that all DNA obey the statistical distribution of an excess in TG/CA/CT dimers, and a deficiency in CG/TA dimer syllables. Furthermore, this distribution is found in coding as well as noncoding regions (and "junk" DNA). The authors point out that the statistical distributions are independent of methylation (CMG converted to TG or CA). The authors also point out that as sense codons (3 base syllables) map into amino acids, the lack of codon randomness derives from the lack of dimer randomness, hence it is to be expected that amino acids do not appear equally distributed in proteins. The authors also point out that the sulfur-containing amino acids Cys and Met are evidence that life started in oceanic geothermal vents.
No actual grammar is explicitly presented in this paper.
"The multiple codes of nucleotide sequences", by E. N. Trifonov, Bulletin of Mathematical Biology, 1989, 51, 4, 417 - 432
The key point in this paper is that there are multiple codes, not just the generally applicable protein codes based upon codons. Multiple codes, not only in DNA, but RNA and proteins as well. The author points out that most of the genome is composed of:
- Intergenic spacer
- Dispersed repetative elements
- Tandem repeated sequences
The three above are named junk DNA, selfish DNA, ignorant DNA, or polite DNA. However, other codes exist, such as:
- General mRNA polyadenylation signals
- TATA-box promotors
- Gene splicing signals
- Species-specific operator sequences
- Phage-specific promotors
- Transcription factor binding sites
- GU and AG intron terminals a, b
- AAUAAA polyadenylation signal a, c
Other codes exist, based upon structural (statistical patterns) or functional (what patterns characterize a given function) criteria. Functional codes will be discussed, but before doing this, the author points out that sequences frequently simultaneously encode for multiple codes by overlapping the same sequence. This is yet another way in which nucleotide sequences differ from human langages. A synthetic, artificial example in written English that exemplifies this is the following. Consider togethernowhere. This is ambiguous, possible interpretations being:
- together nowhere
- together no where
- together now here
- to get her nowhere
- to get her no where
- to get her now here
Translation Framing Code.
H(k)|k=kp=MAX[ Σi=1L–kp{F(i)F(i+k)} ],
where
L is the sequence length,
kp is the distance between the same nucleotides,
F(i) ∈ (0, 1), is the distribution probability function of base A, C, G, T at position "i"
H(k) easily picks out repititions of dinucleotide syllables at multiples of 3 steps. This was found to be explained by frameshift base slipping.
Dinucleotides such as AA and TT repeat along the primary sequence with a period of about 10.5
nucleotides, that appears to correlate with the DNA helix. AA (with Watson-Crick complement TT)
correlates with local DNA helix curvature a (the "roll wedge" and "tilt wedge").
This may be used to map chromatin nucleosome position and spatial conformation. Hence, the
Chromatin code a, where
M(i)=MAX[ Σk=1k=16< Σj=1j=Ln{Fk(i+j-1)Bk(j)} >],
and
Bk(j), j=1..Ln, for the 42=16 dinucleotides,
Fk(i) ∈ (0, 1) is the positional distribution probability function of the
kth dinucleotide sequence from i=1 to L–Ln+1
M(i) has a maximum at DNA folds that correspond to nucleosomes.
Positional correlation directed to amino acid sequences reveals that the distribution of hydrophobic amino acids on the α-helix is not random. This appears to be correlated with the curvature of the α-helices.
Overlapping of two, three, even four messages that span parts of the same stretch of nucleotide sequences show that DNA (and RNA) information is densely packed.
Tandem-repeat sequences in RNA may function as a copy-number to express gene phenotypic enhancers or silencers, thus these codes that appear to be "junk" DNA may indeed not be junk at all! In this context, Trifonov points out that exon codons (triplet code) is spatially separated by the chromatin code contained by introns a, d.
a "DNA AS A LANGUAGE", by E. N. Trifonov, I. Elocin, in
"The Second International Conference on Bioinformatics, Supercomputing and Complex Genome Analysis",
Editors: H. A. Lim, J. W. Fickett, C. R. Cantor, R. J. Robbins, World Scientific, Singapore, 1993,
103 - 110
b "Ovalbumin gene: Evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries", by R. Breathnach, C. Benoist, I. Haras, K. O'Hare, F. Gannon, P. Chambon, Proceedings of the National Academy Of Sciences USA, Oct. 1978, 75, 10, 4853 - 4857
c "3' Non-coding region sequences in eukaryotic messenger RNA", by N. J. Proudfoot, G. G. Brownlee, Nature, Sept. 16 1976, 263, 5574, 211 - 214
d "A general function of noncoding polynucleotide sequences", by E. Zuckerkandle, Molecular Biology Reports, May 22 1981, 7, 1-3, 149 - 158
"CLASSIFICATION AND RELATED METHODS OF DATA ANALYSIS", Edited by H. H. Bock, North Holland,
1988
The paper "NUCLEOTIDE SEQUENCES AS A LANGUAGE: MORPHOLOGICAL CLASSES OF WORDS", by E. N. Trifonov,
in the above book, 57 - 64
The author examines Linear G, genomic texts (languages). Linguistics discussion focuses upon morphology (words), not upon any grammatical analysis. The author states that genomic languages are like human languages in that both are "...one-dimensional arrays of symbols...", thereby excluding higher dimensional languages such as shape grammars or syntactic pattern recognition grammars with the ability to support higher dimensional structures. The author does, however, place heavy emphasis upon semantic considerations such as the "...evolution of matter outside and inside of living organism...", and other areas that will be mentioned. Thus the author's views of semantics in the genome goes well beyond the restrictive views of N. Chomsky (already discussed in the paper by V. A. Ratner). Trifonov points out that the Genomic language (there being only one such language) appears to depart from the similarities to human language in that the genomic "...texts carrying simultaneously several overlapping messages...". Trifonov points out that exons that code proteins typically contain at least three messages. The author of "Emergent Computation: Emphasizing Bioinformatics" has additionally pointed out that human language is rarely simultaneous (as simultaneity interfears with human linguistic transmission and reception), but molecular forces are simultaneously expressed as a language in the case of triple-helix, or quadruplexes, for example.
Trifonov analyzes the genome, examining repetitivenes, morphological classes of genomic words
including "contrast words", "tandem words", and "complementarily symmetrical" words. Significantly,
Tifonov examines "mirror-symmetrical" words. Trifonov points out that these non-stacking,
non-duplexed RNA sequences (which lack complementarity) often appear in RNA bulges and hairpin loops
as sites of RNA-protein binding. Thus Trifonov mentions the RNA Shine-delgarno sequence "GGAGG" and
the mitochondrial RNA "ATTCTTA", as well as the 3'–end processing site in histone genes,
"AGAAAGA". Trifonov mentions the semantically defined "words" such as protein-DNA binding
sites and wedge-like double syllables AA, TT, and possibly CA and TG.
Once again it must be emphasized that Trifonov never goes beyond the linguistic aspects of
morphology, thus never examines grammars.
"Processing of palindromes in neglect dyslexia", by R. C. Shillcock, M. L. Kelly, P. Monaghan, Neuro Report, Sept. 1998, 9, 13, 3081 - 3083
Symmetry is related to human peception of palindromic words as well as visual palindromic structures in the area of neglect dyslexia.
"A Graph Grammar Approach to Artificial Life", by O. Kniemeyer, G. H. Buck-Sorlin, W. Kurth, Artificial Life, 2004, 10, 4, 413 - 431
This paper is an excursion into relational growth grammars (RGGs), considered an extension of parametric Lindenmayer systems. The authors view the languages they descibe purely as models (ie: they do not correspond to any language as an expression of any natural forces, such as molecular attraction). The authors consider interaction with the environment to be significant (globally sensitive and open L systems). The authors note that "A pure rule-based approach is not feasible in all situations. Programmed graph replcement systems solve this problem by supporting additional programming structures." The view of artificial life taken by the authors is apparently derived from Dawkins, and does not appear to be based upon the creation of synthetic molecules (DNA, RNA, or proteins) used in living systems, but rather, a view that is closer to computer game simulations.
"Listening to Viral Tongues: Comparing Viral Trees Using a Stochastic Context-Free Grammar", by A. Rzhetsky, W. M. Fitch, Molecular Biology and Evolution, April 2005, 22, 4, 905 - 913
The authors state that a grammar is a mathematical model that allows generation of a set of strings. The author's views clash violently with the views of N. Chomsky (that languages are effectively limited to human beings, require speech organs, aural organs, psychological aspects, that especially do not apply to the applications the authors apply their systems to), or should have a basis in patterns created by natural laws is not a consideration of the authors, as they view languages strictly as models. The authors limit languages to strings, thereby excluding many Syntactic Pattern Recognition languages (K. S. Fu), as well as shape grammars. The authors require non-terminal symbols (thus excluding most Lindenmayer systems). The authors go on to create a stochastic context-free grammar applied to viral trees (phyogenetic trees). Two probability density functions are used (lognormal and gamma). The authors seem to limit themselves to contexrt-free grammars because of the expense of computation. While the stochastic context-free grammar the authors provide has four non-terminal symbols, no start symbol is explicitly identified (readers must assume this knowledge). Termination of a derivation may use ad-hoc rules such as (depth 3 has been reached, apply a specific substitution rule, in the example provided).
"The generative grammar of the immune system", by N. K. Jerne, Bioscience Reports, 1985, 5, 439 - 451
This lecture contains a lot of very intresting information about the specificity of antibodies, B cells, TH and TK cells. Much information about antibodies (symmetry, constant and variable components), ideotopes, epitopes, etc. Analogies are made to Chomsky type generative grammars, but of so general a nature as to have no or very little value. The author quotes Chomsky requiring a generative grammar to contain a "...central syntactic component..., a phonological component, and a semantic component.". The author points out that Chomsky feels that "...certain deep, universal features of this competence are innate characteristics of the human brain." Of course, such a linguistic form of racism has yet to be demonstrated, and is precisely what the author of "Emergent Computation: Emphasizing Bioinformatics" is opposed to (molecular language does not appear to require a phonetic, aural, or even a anthropomorphic-semantic aspect). The author of "Emergent Computation: Emphasizing Bioinformatics" does not require innate undemonstrated aspects of grammar in human beings, the more so as there are so many different and inequivalent systems of grammars. Jerne ends his lecture with the view that "... an inheritable capability to learn any language means that it must somehow be encoded in the DNA of our chromosomes. Should this hypothesis one day be verified, then linguistics would become a branch of biology." Thus Jerne would seem to feel that DNA molecular languages (in opposition to Chomsky's speculative prejudices) are possible. However, synthetic bases used in DNA and RNA, as well as synthetic polypeptides, etc. suggest that any linguistic view of DNA, RNA, and proteins extends well beyond biology.
"Hydraulic and channel characteristics of selected streams in the Kantishna Hills area, Denali Nataional Park and Preserve, Alaska", by J. L. Van Maanen, G. L. Solin, Dept. of the Interior, U.S. Geological Survey, 1982-84
Alluvial River Channels, formed under the forces of gravity in river deltas, as rivers flow down mountain slopes, etc. Rivers carry sediments, with specific rates and distances. River beds move or meander (on a flood plain). Rivers often have a braided morphology, but often have a tree-like morphology, especially when flowing down hillslopes. The more predictable the morphology (due to a variety of forces expressed as patterns), the greater the likelihood of linguistic patterns.
Shape Grammar for Alluvial River Channels
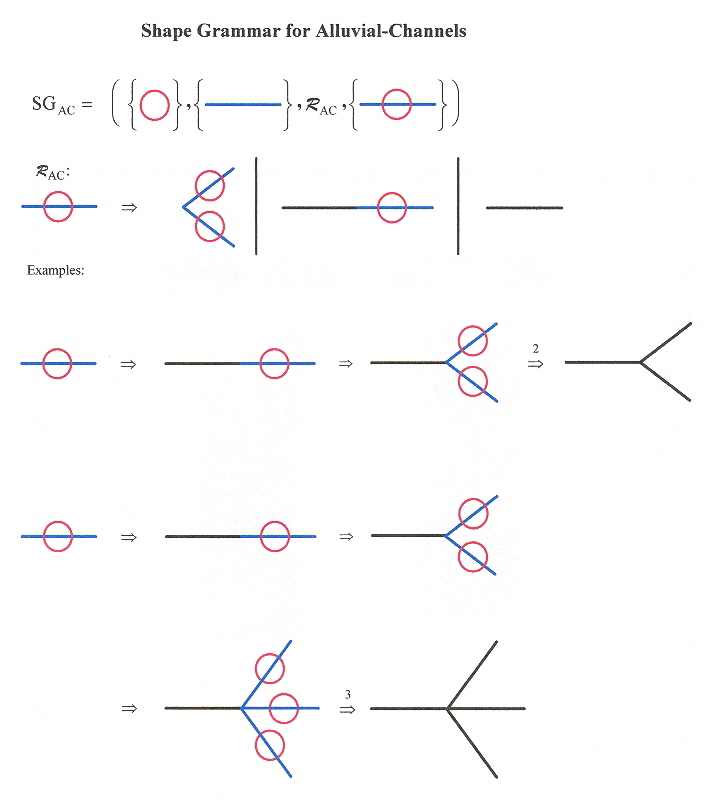
Example of an Alluvial River Channel

"Knowledge Management through Knowledge Engineering", by A. Preece, A. Flett, D. Sleeman, D. Curry, N. Meany, P. Perry, IEEE Intelligent Systems, 2001, 36 - 43
The authors attempt to define a "grammar" for geology applications. Their putative "grammar" appears to be as follows (slightly modified):
G = ( VN, VT, P, <formation> ), where:
VN = { <formation>, <lithology>, <lithology-depth>, <lithology-length>,
<rock>, <rock-type>, <rock-hardness> }
VT = { shale, clay, chalk, granite, other, very-soft, soft, medium, hard, very-hard }
The set of production rule "P" follow:
<formation> ⇒ <lithology> <formation> | <lithology>
<lithology> ⇒ <rock> <lithology-depth> | <rock> <lithology-depth>
<lithology-length>
<rock> ⇒ <rock-type> <rock-hardness>
<rock-type> ⇒ shale | clay | chalk | granite | other
<rock-hardness> ⇒ very-soft | soft | medium | hard | very-hard
<lithology-depth> and <lithology-length> being undefined, except as numeric values as it is
deemed that these non-terminal symbols are too difficult to define with a "grammar" (hence this putative
"grammar" isn't a grammar at all). It would probably be better to define the twenty-five terminal strings generated by
<rock> ⇒ <rock-type> <rock-hardness> as <rock> ⇒ <rock-hardness>
<rock-type>, as most people would prefer the English 'soft clay' to 'clay soft', but this is a minor
point.
This is an important paper in that the authors do extend linguistic methods to the study of Geology.
There are very few papers that extend linguistics to geology, meterology, oceanography. However, there are
linguists (such as N. Chomsky) that append requirements including speech organs and aural organs to
support phonetics, and psychological states to language in general. Such anthropomorphic aspects
thereby becoming a requirement for linguistics to be used (except in a very limited way, only as a
mathematical tool). Such a view of linguistics imposes an extremely limited, anthropomorphic view of
semantics monolithically across the totality of the study of linguistics.
The authors of this paper about geology totally ignore (or are unaware of) the fact that a new, non
anthropomorphic semantics is being invoked, thus expanding the study of linguistics!
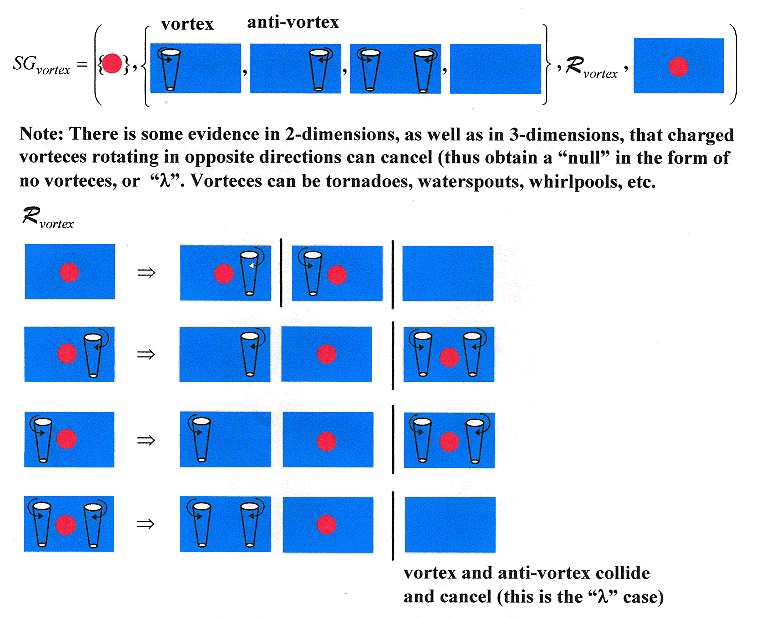
"Vorteces in the XY model", by Evgeny Demidov (2004), Rong Feng Sun (Clark University), internet search under "vortex antivortex collapse".
Vorteces and anti-vorteces, especially in 2-dimensions, but also in 3-dimensions can collide and collapse into a null state (λ). A Shape language for such vorteces appears below. Note that the reference refers to charged vorteces. Vorteces include tornadoes, waterspouts, and whirlpools, and other possibilities, such as plasmas. Vorteces with their anti-vorteces may cancel in the three-dimensional case as in three dimensions, a vortex may be viewed as two-dimensional planes stacked up vertically. In the three-dimensional case, while vortex/anti-vortex pairs can cancel in each two-dimensional plane, it is also possible for a vortex in one plane to cancel with its anti-vortex in an adjacent plane.
A Vortex Language
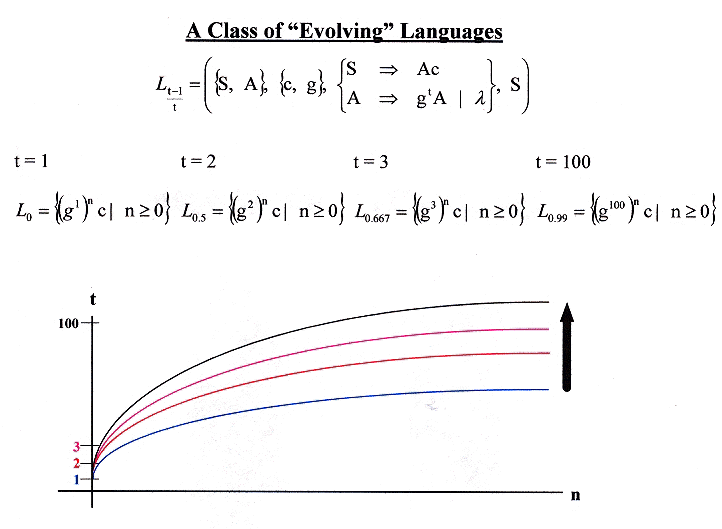
Evolution of Languages

Evolution of Languages