Frame Shift type Codon usage
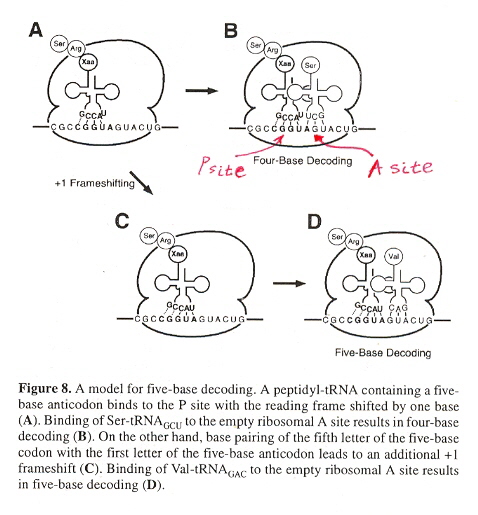
"Five-base codons for incorporation of nonnatural amino acids into proteins', by T. Hohsaka, Y. Ashizuka, H. Murakami, M. Sisido, Nucleic Acids Research, 2001, 29, 17, 3646 - 3651
Examining Figure 8 (below, for a review of Frame-Sense Hopping and Sliding, see "Hopping and Slidding" in Hopping.html. We get an idea what happens at the ribosome when there are mixed-length anticodons. When a longer anticodon is encountered, it is partially accomodated at the P site, the next tRNA anticodon being read at the A site. To fully accomodate the longer anticodon, a +1 or +2 slip frameshift at the P site takes place, followed by the next tRNA anticodon at the A site.
"Exploring the Limits of Codon and Anticodon Size", by J. C. Anderson, T. J. Magliery, P. G. Schultz, Chemistry & Biology, Feb. 2002, 9, 237 - 244
N-length codons prefer N+4-length anticodon loops (or N+3-length anticodon loops):
Codon tRNA loop anticodon2 6
3 7
4 8
5 9
A 1 nt base deletion at the anticodon implies a – 1 frameshift (slip)
A 1 nt base insertion at the anticodon implies a + 1 frameshift (slip)
A 2 nt base deletion at the anticodon implies a – 2 frameshift (slip)
A 2 nt base insertion at the anticodon implies a + 2 frameshift (slip)
A 3 nt base deletion at the anticodon implies a – 3 frameshift (slip)
A 3 nt base insertion at the anticodon implies a + 3 frameshift (slip)
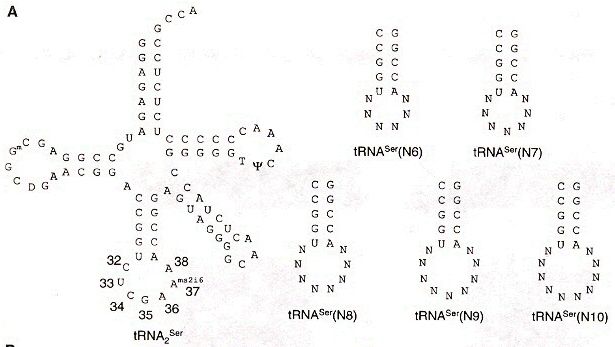


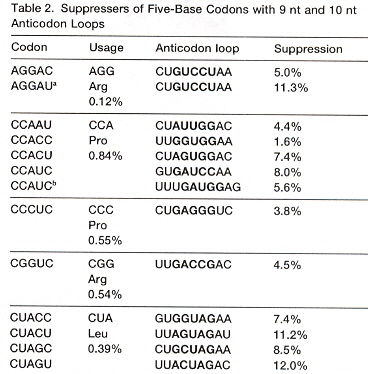
If there are insufficient bases in the anticodon loop, delete a base from the 5' end of the anticodon, for each base to be deleted (see the table cross-referencing Nucleotides in anticodon loop with Decoding patterns, above ).
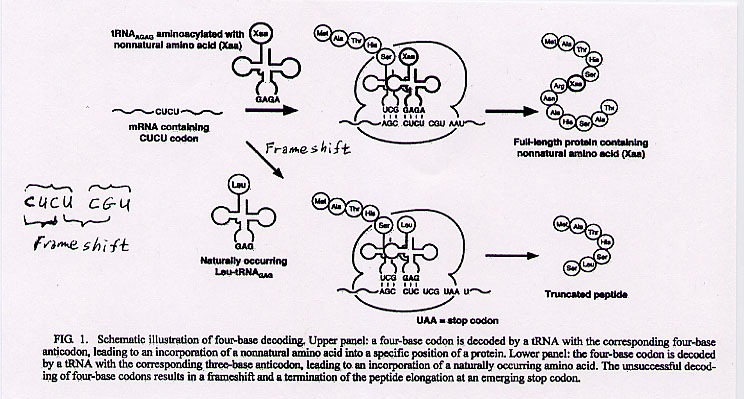
"Incorporation of Nonnatural Amino Acids into Proteins by Using Various Four-Base Codons in an Escherichia coli in Vitro Translation System", by T. Hohsaka, Y. Ashizuka, H. Taira, H. Murakami, M. Sisido, Biochemistry, 2001, 40, 11060 - 11064
Efficient incorporation of two nonnatural amino acids into a single protein using codons composed of four bases. More than two nonnatural amino acids may be incorporated into proteins, but efficiencies (rate of incorporation) are not high.
"Expanding the Genetic Code in a Mammalian Cell Line by the Introduction of Four-Base Codon/Anticodon Pairs", by M. Taki, J. Matushita, M. Sisido, ChemBioChem, 2006, 7, 425 - 428
The following orthogonal sets of four-base codons have been used in E. coli:
- (CGGG, AGGU)
- (CGGG, GGGU)
- (CGGG, GCCC)
New sets of four-based codons that would work in Mammalian cell lines are needed. Specifically, four-base codons of the amber stop-codon form UAGN where N ∈ {A, U, G, C}, with anticodon NCUA. tRNATyrncua was used, where N ∈ {A, U, G, C}. The results found were that four-base codons UAGG/ccua and CUAU/auag could be used to insert non-natural amino acids in mammalian polypeptide chains. Note that five-base codons were sought as well, but none of these functioned at a very high level.
"Efficient Synthesis of Nonnatural Mutants in Escherichia coli S30 in vitro Protein Synthesizing System", by K. Yamanaka, H. Nakata, T. Hohsaka, M. Sisido, Journal of Bioscience and Bioengineering, 2004, 97, 6, 395 - 399
Using the 4-base codon system to insert nonnatural amino acids into proteins, lower than expected yields in in vitro systems were observed. A likely cause of these low yields was the competing production of cyclic tRNA.
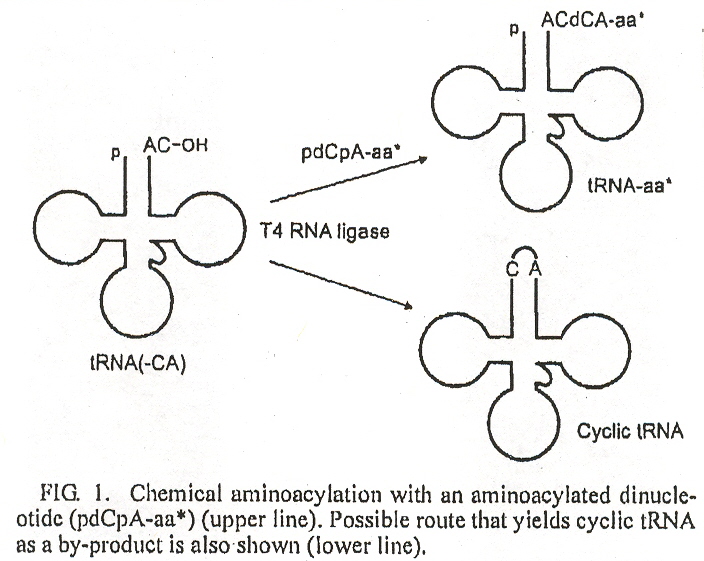
Read-through type Codon usage
"An Expanded Eukaryotic Genetic Code", J. W. Chin, T. A. Cropp, C. Anderson, M. Mukherji, Z. Zhang, P. G. Schultz, Science, August 15 2003, 301, 964 - 967
Incorporation of unnatural (alien) amino acids in response to the nonsense codon TAG in vitro and in vivo.
"A Genetically Encoded Photocaged Amino Acid", by N. Wu, A. Deiters, T. A. Cropp, D. King, P. G. Schultz, Journal of the American Chemical Society, Nov. 10 2004, 126, 44, 14306 - 14307
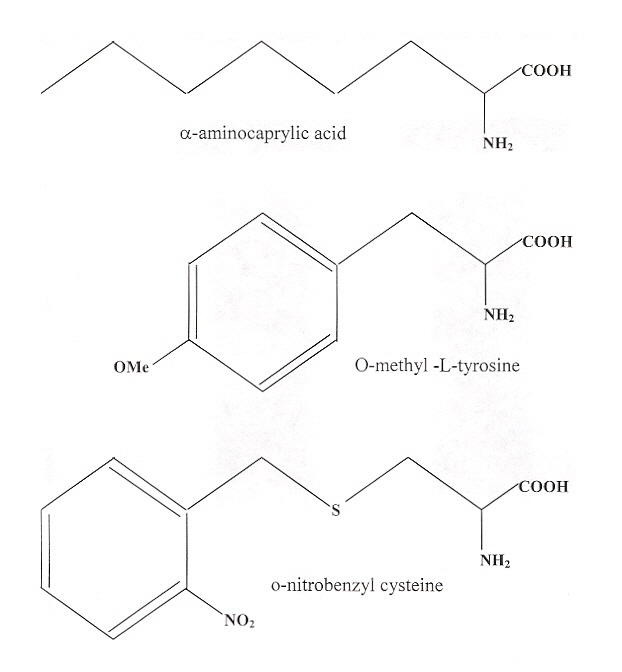
A new pair of Escherichia coli tRNALeu/leucyl-tRNA-synthetase are used to place unnatural amino acids when the amber nonsense codon is encontered (as described in Chapter 3 of "Emergent Computation: Emphasizing Bioinformatics"). The nonnatural amino acids are in the figure below. Furthermore, o-nitrobenzyl cysteine may be used to photoregulate cysteine protease caspase 3.
"Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion", by T. Kobayashi, K. Sakamoto, T. Takimura, R. Sekine, V. P. Kelley, K. Kamata, S. Nishimura, S. Yokoyama, Proceedings of the National Academy of Sciences of the U.S., Feb. 1, 2005, 102, 5, 1366 - 1371
Alloproteins (proteins containing nonnatural amino acids) with site-specific nonnatural amino acids engineered by expanding the natural genetic code using TyrRS (tyrosyl-tRNA synthetase) discussed in Chapter 3 of "Emergent Computation: Emphasizing Bioinformatics". The effect is to substitute the nonnaturally occurring 3-iodo-L-tyrosine in lieu of L-tyrosine both in vitro and in vivo in response to the amber codon. The ultimate object is to produce alloproteins used as molecular switches (to signal pathways), photocrosslinkers, probes incorporating fluorescent labels, or heavy-atom alloproteins used in x-ray structural studies.
"Synthesis and sequence optimization of GFP mutants containing aromatic non-natural amino acids at the Tyr66 position", D. Kajihara, T. Hohsaka, M. Sisido, Protein Engineering, Design & Selection, May 31, 2005, 18, 6, 273-278
Random mutation around non-natural amino acids is a useful strategy in order to improve protein functions.
- Fifteen aromatic non-natural amino acids are studied:
- L-1-naphthylalanine,
- L-2-naphthylalanine,
- L-p-bipphenylalanine,
- L-2-anthrylalanine,
- L-2-pyrenylalanine,
- L-p-nitrophenylalanine,
- L-p_dimethylaminophenylalanine,
- L-3-(9-ethylcarbazolyl)alanine,
- L-azatryptophan,
- L-kynurenine,
- L-p-phenylazophenylalanine,
- L-p-benzoylphenylalanine,
- L-2-anthraquinonylalanine,
- L-p-aminophenylalanine,
- L-O-methylthyrosine,
"Four-Base Codon-Mediated Incorporation of Nonnatural Amino Acids into Proteins in a Eukaryotic Cell-Free Translation System", by H. Taira, M. Fukushima, T. Hohsaka, M. Sisido, Journal of Bioscience and Bioengineering, May 2005, 99, 5, 473 - 476
The expansion of the 64 codon set including an expanded 65th anti-codon that transcribed a non-natural amino acid was discussed in Chaper 3 of "Emergent Computation: Emphasisizing Bioinformatics". In this exciting paper, an alternative is used: the use of mixed 3- and 4-base codons, using frameshift suppression. Obviously, with a 4-based codon set and 4 bases, there are 44=256 possible codons (not using nonnatural bases). These 4-base codons were used to successfully introduce nonnatural amino acids into proteins. CGGU, CGCU, CCCU, CUCU, CUCA, and GGGU efficiently functioned with their anti-codons. The 4-base codons AGGU, AGAU, CGAU, UUGU, UCGU, and ACGU were not decoded. The standard 3-base amber (nonsense) codon UAG, and the opal codon UGA worked efficiently, while the 4-base stop codons UAGU, UGAU, and UAAU were inefficient. tRNAs were aminoacylated and charged with nonnatural amino acids and these tRNAs had 4-based codons. These codons worked efficiently in Escherichia coli, but not so well in rabbits.
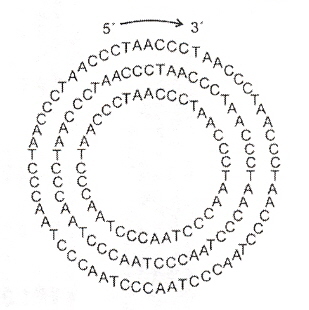
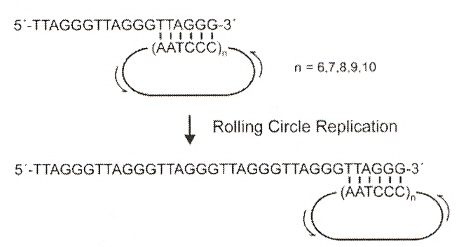
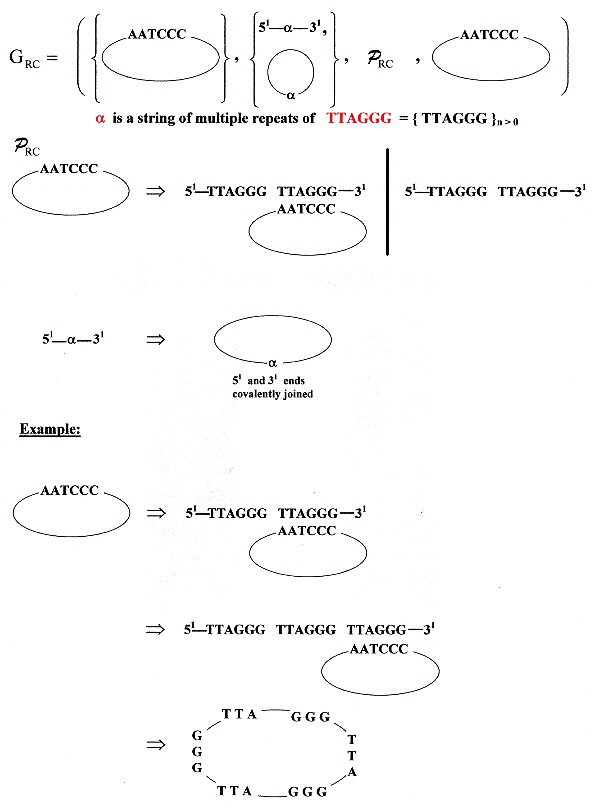
Rolling Circle Repeats of Telomeric Sequences
"Small circular DNAs for synthesis of the human telomere repeat: varied sizes, strctures and telomere-encoding activities", by J. S. Hartig, E. T. Kool, Nucleic Acids Research, Nov. 1 2004, 32, 19, e152, 1 - 6
Circular DNA oligonucleotides composed of the human telomere repeat (CCCTAA)n have been constructed. These range in size from 36 to 60 oligonucleotides. Using 18mer telomeric primers and DNA polymerases, these circular oligonucleotides act as rolling circle templates to synthesize telomeric repeats containing in excess of 1000 nucleotides.
|
Rolling Circle Oligonucleotides
|
Rolling Circle Replication
|
Rolling Circle Replication Grammar
Boolean Algebra of Codons
"A genetic code Boolean structure. I. The meaning of Boolean deductions", by R. Sánchez, E. Morgado, R. Grau, Bulletin of Mathematical Biology, 2005, 67, 1 - 14
A Boolean Algebra of codons (and its dual) is created where bases are associated with their triples: U → UUU, C → CCC, G → GGG, A → AAA, and Watson-Crick complements maintained: CCC corresponding to GGG, and UUU (or TTT) corresponding to AAA. The Boolean algebra is isomorphic to ( (Z2)6, ∨, ∧ ), using a binary encoding as follows:
G ↔ 00, A ↔ 01, U ↔ 10, C ↔ 11, the following is obtained (where ∨ corresponds to "or" and ∧ corresponds to "and", negation ¬ inverts 0 and 1):
thus CAG &or AUC ↔ 110100 ∨ 011011 = 111111 ↔ CCC and ¬[ CAG ∧ AUC ] ↔ ¬[ 110100 ∧ 011011 ] = ¬[ 010000 ] = 101111 ↔ UCC
Note that a codon in the 5' → 3' direction (in the primal algebra) is matched by the anticodon in the 3' → 5' direction (in the dual algebra).
Thus if X1Y1Z1 → X2Y2Z2 in the primal algebra, then X2Y2Z2 → X1Y1Z1 in the dual algebra.
Also note that if X1Y1Z1 → X2Y2Z2, then ¬ (X1Y1Z1) ∨ (X2Y2Z2) = 1 (in the algebra).
For example, AUG → CUG ≡ ¬ (AUG) &or (CUG) = UAC ∨ CUG = CCC = (111111) = 1 in the primal algebra (use the tables below).
|
|
|
Genetic mutations associated with drug resistance HIV protease and beta-globin variants are then correlated to codon mutations.
"A genetic code Boolean structure. II. The genetic information system as a Boolean information system", by R. Sánchez, R. Grau, Bulletin of Mathematical Biology, September, 2005, 67, 1017 - 1029
If indices j and k vary over 1 .. 20 (to encode the common amino acids), and index i designates the number of codons, then the value of information is as follows: ni= ( j, k ). If codon X2Y2Z2 is deduced from codon X1Y1Z1 or X1Y1Z1 &rarr X2Y2Z2. Amino acids M (methionine) has one codon "AUG" and Y (tyrosine) has two codons "UAU" and "UAC".
Thus note that GGG → AUG as ¬ (GGG) ∨ (AUG) = (CCC) ∨ (AUG) = (CCC) = (111111) and note that GGG → UAU as ¬ (GGG) ∨ (UAU) = (CCC) ∨ (UAU) = (CCC) = (111111), thus n1(AUG, UAU) = 1 (AUG and UAU are both deduced from one codon).
also note that ( CCU ∧ CCC ) → AUG as ¬ (CCU ∧ CCC) ∨ (AUG) = ¬ (CCU) ∨ ¬ (CCC) ∨ (AUG) = (GGA) ∨ (GGG) ∨ (AUG) = (GGG) = (111111) in the dual.
also note that ( CCU ∧ CCC ) → UAU as ¬ (CCU ∧ CCC) ∨ (UAU) = ¬ (CCU) ∨ ¬ (CCC) ∨ (UAU) = (GGA) ∨ (GGG) ∨ (AUG) = (GGG) = (111111) in the dual.
thus n2(AUG, UAU) = 2 (AUG and UAU are both deduced from two codons).
Similarly, CCC → AUG and CCC → UAC, thus in the dual algebra we obtain n2(AUG, UAC) = 1 (AUG and UAC are both deduced from one codon).
The final result is that n(M, Y) = n1 ( AUG &and UAU ) + n1 ( AUG, UAC) + n2 ( AUG &and UAU ) + n2 ( AUG &and UAC ) = 5 (indicating redundancy).
Finally, the information value of amino acid (given redundancy) is expressed by the formula:
Vi= – log4 [ { ∑ j=120> n(i,j) } / N] , where N=∑i=120 ∑j=120 n(i,j)
Back to Top