DNA/cDNA, RNA Nucleic Acid Bases
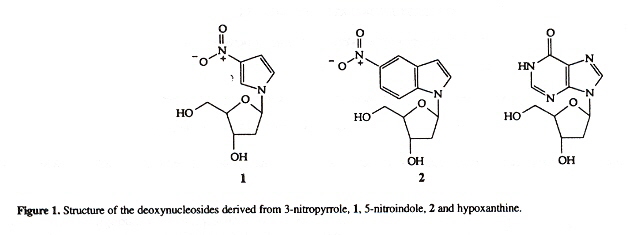
Universal bases are bases that pair with all of the natural bases (A, C, G, T) without discriminationa. Degenerate bases show marked preferences for pairing with either pyrimidines or purines. There are few examples of truly universal bases. Three possibilities are hypoxanthine, 3-nitropyrrole, and 5-nitroindoleb.
a "5-Nitroindole as an universal base analogue", by D. Loakes, D. M. Brown, Nucleic Acids Research, 1994, 22, 20, 4039 - 4043
b "3-Nitropyrrole and 5-Nitroindole as an universal bases in primers for DNA sequencing and PCR", by D. Loakes, D. M. Brown, S. Linde, F. Hill, Nucleic Acids Research, 1995, 23, 13, 2361 - 2366
3-nitropyrrole, 5-nitroindole, hypoxanthine
Stability of heteroduplexes
"A 68-Nucleotide Sequence within the 3' Noncoding Region of Simian Hemorrhagic Fever Virus Negative-Strand RNA Binds to Four MA104 Cell Proteins", by Y-K Hwang, M. A. Brinton, Journal of Virology, Jan. 12 1998, 4341 - 4351
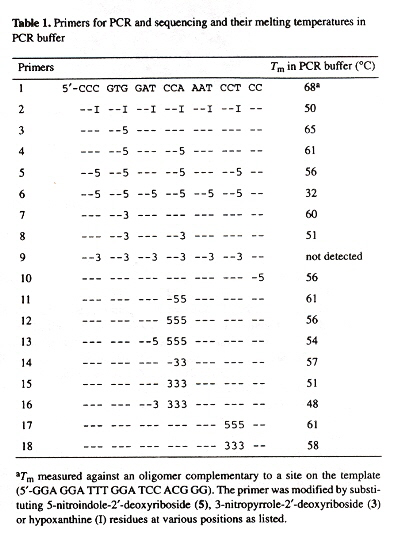
Simian Hemorrhagic Fever Virus (SHFV) genome encodes a nonstructural polyprotein at its 5' end, and structural proteins at its 3' end (where mRNAs are produced during replication). These mRNAs contain a 5' leader sequence identical to the one at the 5' end of the genome (about 209 nucleotides in length or the entire noncoding region or NCR after the leader sequence). After the NCR, there are overlapping open reading frames or ORFs. There is evidence that host proteins are involved in RNA virus replication. Four cellular proteins in MA104 cells bind to the 3' end of SHFV RNA. The minimal binding region is at nucleotides 117 to 184, 68 nucleotides in length). The SHFV binding region was determined by using different combinations of eleven primer sequences. cDNA templates were created by PCR and used for in vitro RNA transcription.
The predicted secondary structure of SHFV RNA (using the MFold software, version 9.0)
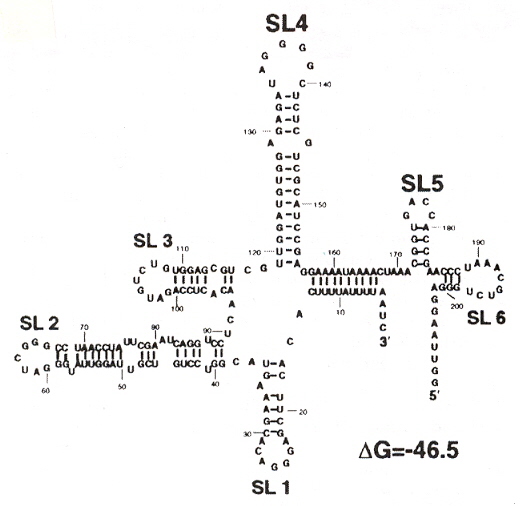
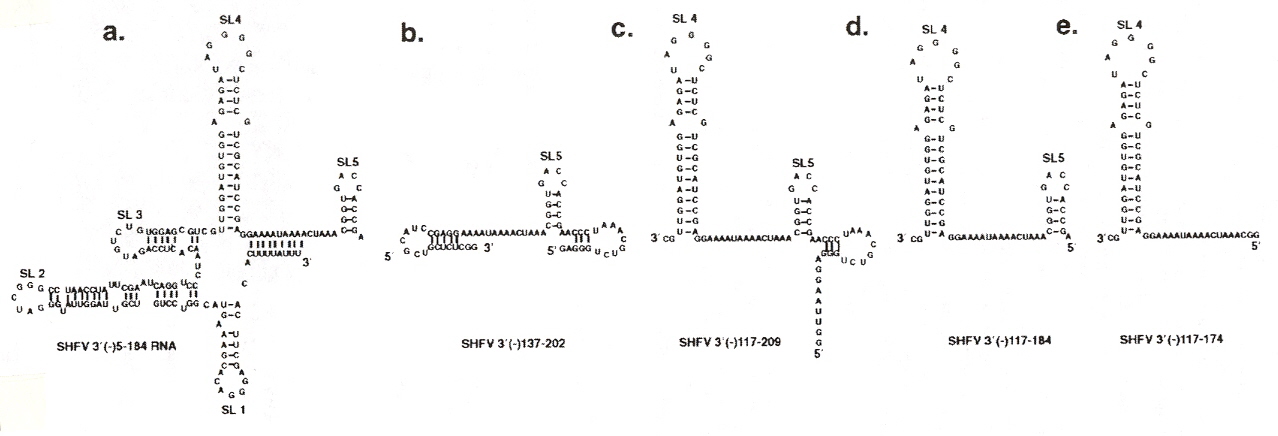
Using the eleven primer sequences that still conserve binding to the four cellular proteins, it was determined that stem loops 4 and 5 (SL4 and SL5) are required, and that the the minimal nucletide sequences for the binding sites are between 117 and 184.
Localization of SHFV RNA Folds for Different Nucleotide Binding Sequences
"SURVEY and SUMMARY The non-Watson-Crick base pairs and their associated isostericity matrices", N.B. Leintis, J. Stombaugh, E. Westhof, Nucleic Acids Research, 2002, 30, 16, 3497 - 3531
-
Collected observations of non-Watson-Crick base pairs in RNA. Twelve Cis/Trans pairings, using either:
- Watson-Crick/Watson-Crick,
- Watson-Crick/Hoogstein,
- Hoogstein/Hoogstein,
- Watson-Crick/Sugar-Edge,
- Hoogstein/Sugar-Edge,
- Sugar-Edge/Sugar-Edge bonding,
with attention to RNA secondary structures.
"Prolegomena to Future Experimental Efforts on Genetic Code Engineering by Expanding Its Amino Acid Repertoire", by N. Budisa, Angewandte Chemie International Edition, 2004, 43, 6426 - 6463
Although this paper is but a summary of results found concerning the expansion of amino acids typically not naturally found in proteins, this paper is an outstanding and very useful paper. Early examples of engineered "alien" type life (with unnatural amino acid proteins) are mentioned a, showing that efforts to engineer organisms that incorporate non-natural amino acids have a a longer history than many are aware of (hence the repeated claims to be the "first to ...").
Sections 4 and 5 of this paper are most interesting. There is simply too much valuable information to summarize here, thus it is recommend that this paper be read in its entirety. A couple of figures from the paper will be reproduced. The importance of selective pressure methods has been stressed. Failures due to the inability to appreciate evolutionary-developed interactions between proteins as well as DNA, RNA, etc. have also been stressed.
This paper includes supression methods. Nonsense codon read-throughb, as well as frameshift read-throughc.
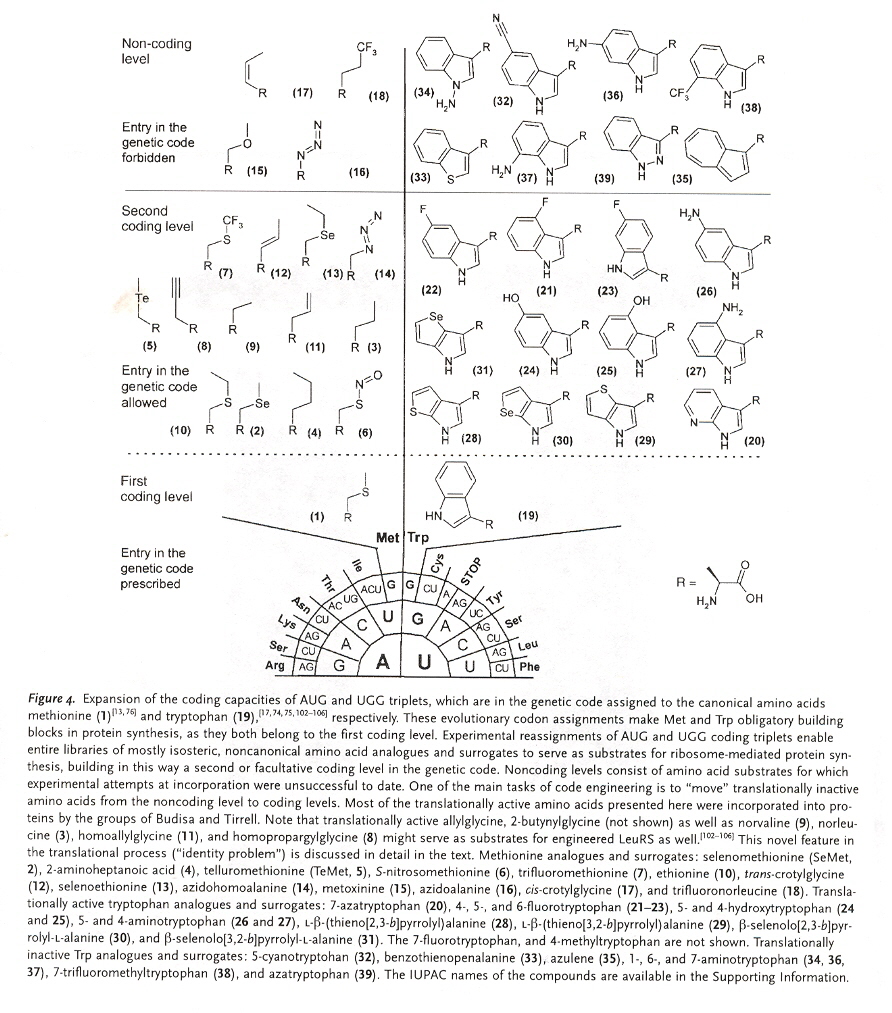
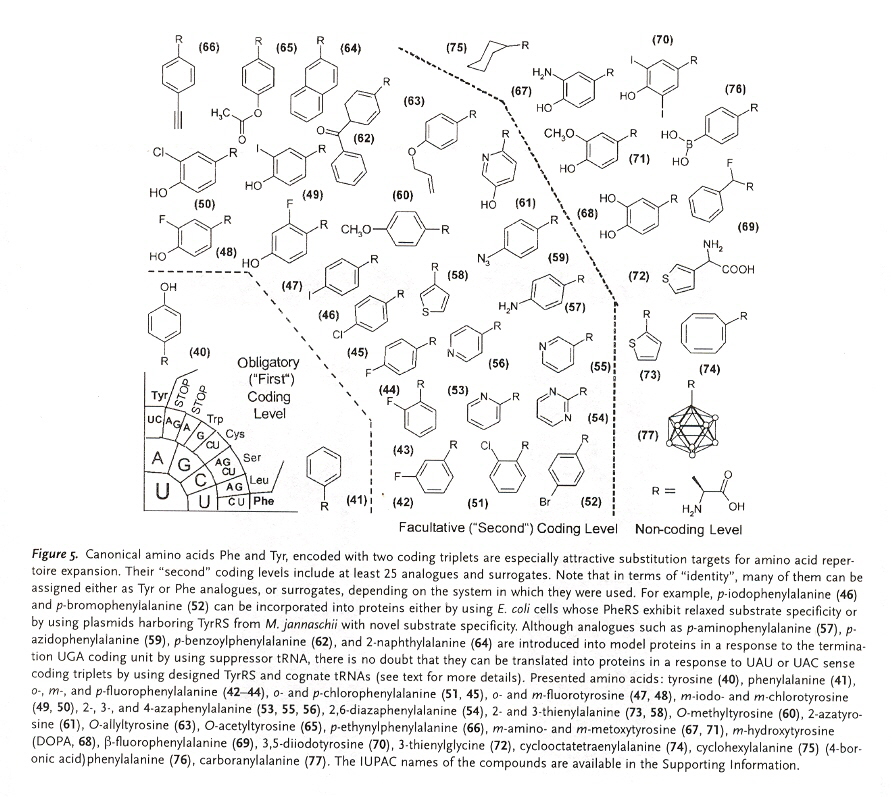
- J. T. F. Wong, Proceedings of the National Academy of Sciences of the U.S.A., 1983, 80, 6303
- D. Mendel, V. W. Cornish, P. G. Schultz, Annual Review of Biophysics and Biomolecular Structure, 1995, 24, 435
- T. Hohsaka, M. Sisido, Current Opinion in Chemistry and Biology, 2002, 6, 809
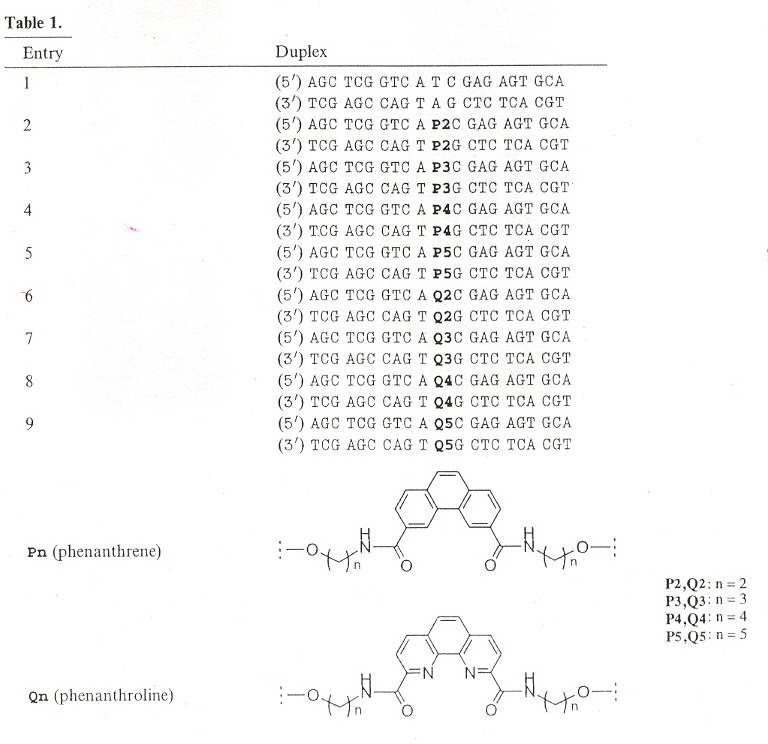
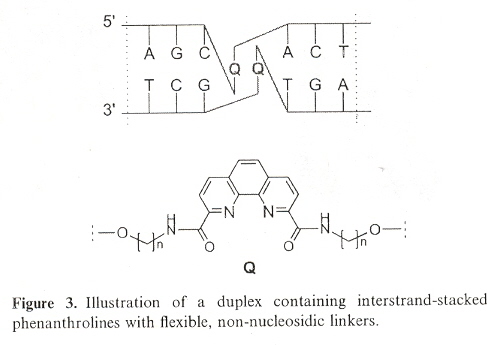
"DNA containing phenanthroline- and phenanthrene-derived, non-nucleosidic base surrogates", by S. M. Langenegger and R. Häner, Tetrahedron Letters, Dec. 6 2004, 45, 50, 9273 - 9276
Non-nucleoside phenanthroline derivatives and phenanthrene derivatives are introduced into double-stranded oligodeoxynucleotide polymers to form stable hybrids in which there is interstrand stacking of the non-nucleosides.
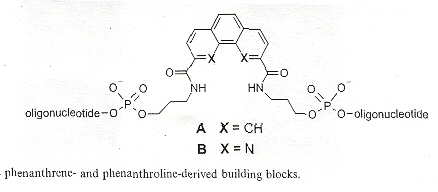
"Detection of single base mismatches and abasic sites using phenanthridium as an artificial base and charge donor", by L. Valis, N. Amann, H-A. Wagenknecht, Organic and Biomolecular Chemistry, 2005, 3, 36 - 38
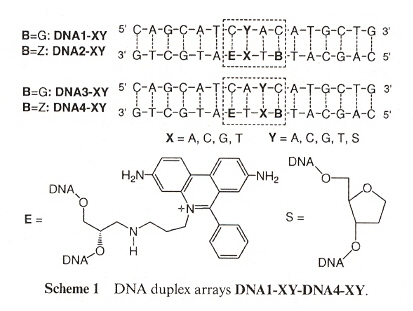
Phenanthridium acts not only as an artificial DNA base, but may also take part as a fluorescent probe. Ethidium (E) also is a chromophore, acting as a photoinduced charge donor. 7-deazaguanine (Z) acts as a charge acceptor. Both E and Z act as DNA bases. Examining Scheme 1 below, we must underatand that Y and X are coupled, where Y = A, C, G, T, S (S means abasic), and X = A, C, G, T. Thus Y/X coupling is one of: (A, A), (A, C), (A, G), (A, T), (C, A), (C, C), (C, G), (C, T), (G, A), (G, C), (G, G), (G, T), (T, A), (T, C), (T, G), (T, T), (S, A), (S, C), (S, G), (S, T). B can either be G or Z, thus a C/B coupling means (C, G), (C, Z). The result is that E will fluoresce if there is a mismatch or an abasic coupling.
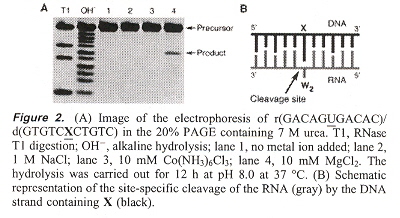
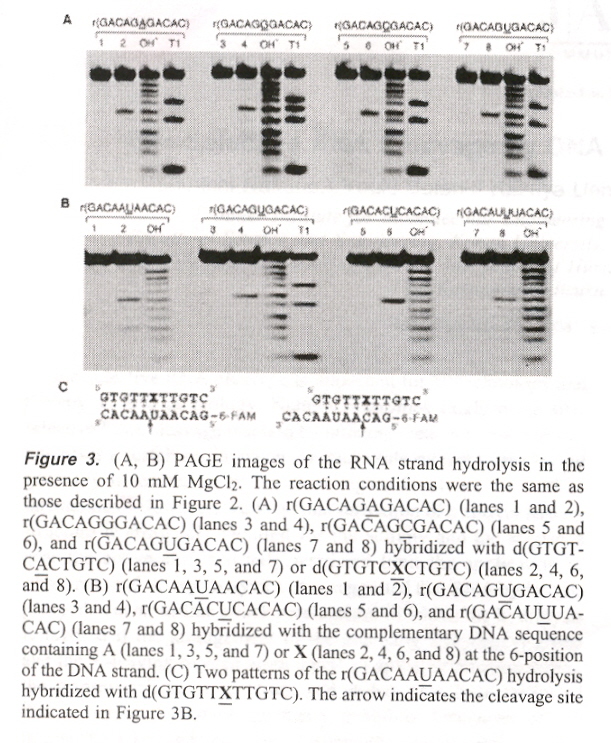
"Site-Selective RNA Cleavage by DNA Bearing a Base Pair-Mimic Nucleoside", by S-I. Nakano, Y. Uotani, K. Uenishi, M. Fuhii, N. Sugimoto, Journal of the American Chemical Society, Jan. 19 2005, 127, 2, 518 - 519
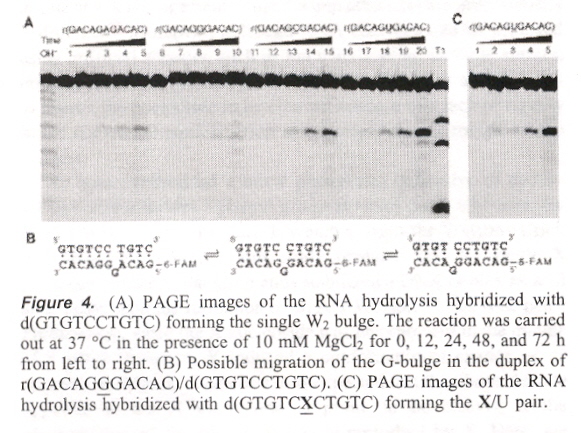
If X = N6 - (N1 - phenylcarbamoyl) - 21 - deoxyadenosine, then {X, A, abasic} on DNA, may be paired with {A, C, G, U} on RNA in a double-helix DNA/RNA hybrid. When this is so, X (in the presence of MgCl2) will cleave RNA (at such sites), except that in the abasic DNA/G-bulge case, pairs shift (rearrange) towards the DNA 3' side.
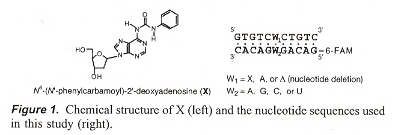
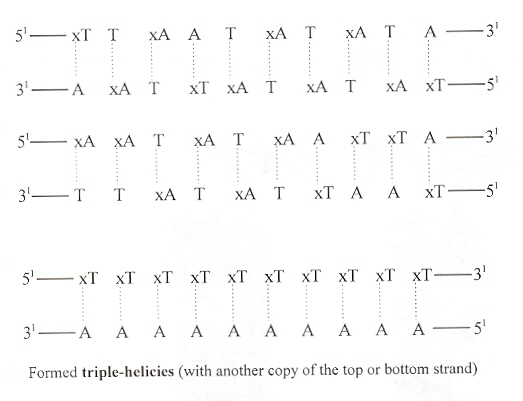
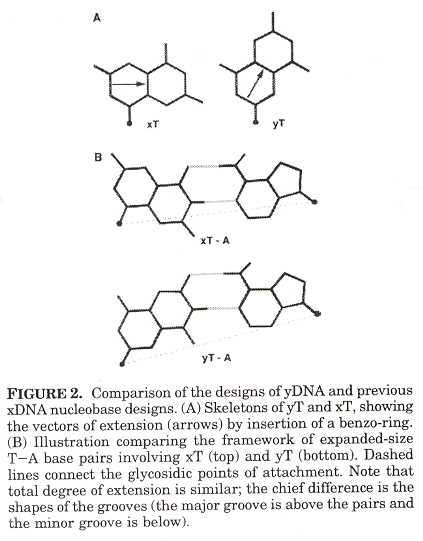
"Helix-Forming Properties of Size-Expanded DNA, an Alternative Four-Base Genetic Form", by H. Liu, J. Gao, E. T. Kool, Journal of the American Chemical Society, Feb. 9 2005, 127, 5, 1396 - 1402
Exploration of T/xA and xT/A expanded base DNA (xT/xA pairs are too large for the xDNA helix). It is particularly interesting that a pyrimidine-rich strand of xDNA formed a triple-helix.
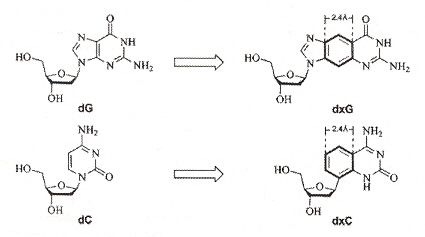
"Size-Expanded Analogues of dG and dC: Synthesis and Pairing Properties in DNA", by H. Liu, J. Gao, E. T. Kool, Journal of Organic Chemistry", Jan. 19 2005, 70, 2, 639 - 647
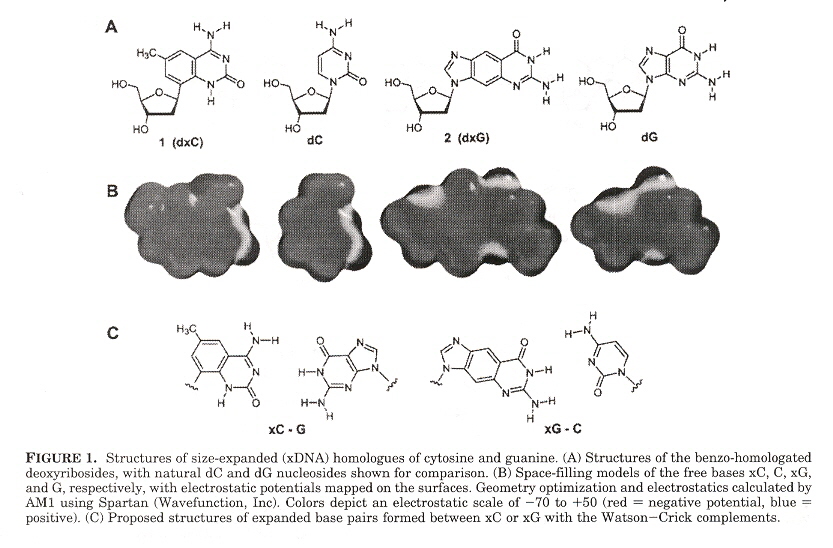
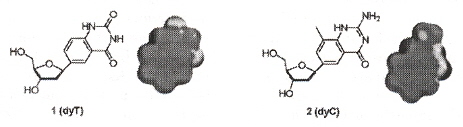
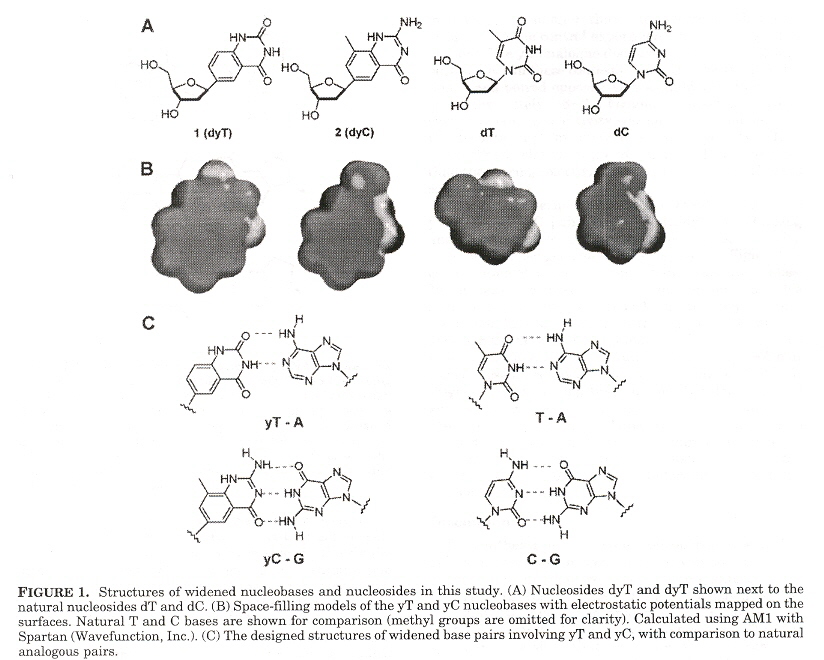
"Novel Benzopyrimidines as Widened Analogues of DNA bases", by A. F. Lee, E. T. Kool,Journal of Organic Chemistry", Jan. 7 2005, 70, 1, 132 - 140
Expanded DNA analogues used to test the stability of an expanded DNA were synthesized. The expanded DNA base analogues have a benzene ring added which also greatly increases the flourescence of these new base analogues. Two sets of expanded base analogues were created, along different axis (see the fifth figure) of the added benzene ring. These are referred to as xDNA and yDNA. When mixing standard bases and these expanded DNA bases, stability decreases, but stability is very good when xDNA (or yDNA) double helix are unmixed.
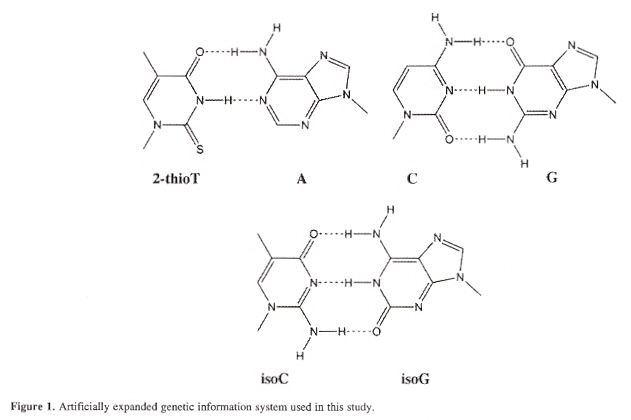
"The use of thymidine analogs to improve the replication of an extra DNA base pair: a synthetic biological system", by A. M. Sismour, S. A. Benner, Nucleic Acids Research, Sept. 28 2005, 33, 17, 5640 - 5646
Considering the isoG/isoC base pair, isoguanine pairs with thymine causing base-pair matching infidelities. It is proposed to pair A with 2-thioT, the supply coming not from TTP, but from 2-thioTTP. The new base alphabet would then be: A/2-thioT, C/G, isoC/isoG. PCR fidelity is then 93%.
Thymidine analog DNA base alphabet
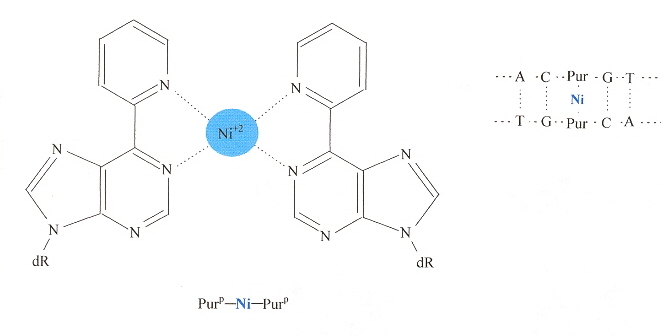
"A Purine-like Nickel(II) Base Pair for DNA", by C. Switzer, S. Sinha, P. H. Kim, B. D. Heuberger, Angewandte Chemie International Edition, Feb. 25 2005, 44, 10, 1529 - 1532
Purp is derived from adenine by replacing the 6-amino group (top of page 21 in "Emergent Computation: Emphasizing Bioinformatics) by a pyridyl group. The result allows two Purp to form a complex with Nickel(II) that can act like a self-complementing base pair in a duplex dDNA.
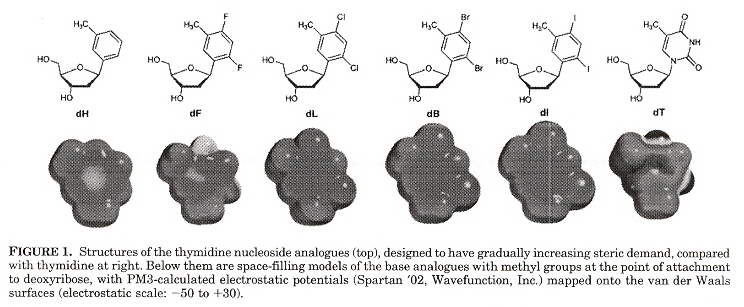
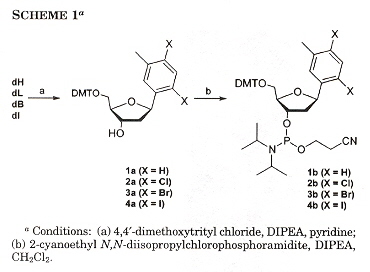
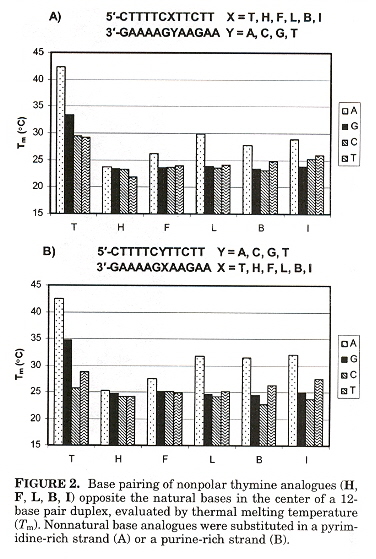
"A Series of Nonpolar thymidine Analogues of Increasing Size: DNA Base Pairing and Stacking Properties", by T. W. Kim, E. T. Kool, Journal of Organic Chemistry, March 18 2005, 70, 6, 2048 - 2053
In other papers by these investigators, synthetic DNA bases have varied in shape while attempting to keep size as constant as possible. In this paper, the size of synthetic bases is gradually increased while keeping the shape as constant as possible. Thymidine analogues especially with halogens have been examined. These analogues are referred to as dF (Fluorine), dL (Chlorine), dB (Bromine), dI (Iodine), while dT refers to thymidine. Results found were that all molecules were destabilizing compared to T. Stability tended to increase as the analogue increased in size. Paired against each other, dH/dH and dF/dH were most destabilizing. dL, dB, dI in the pyrimidine strand and dB, dI in the purine strand were less destabilizing. dL/dI, dB/dI, dI/dI were found to be as stable as the natural T/A pair. Thus dI/dI was most stable, dH/dH least stable.
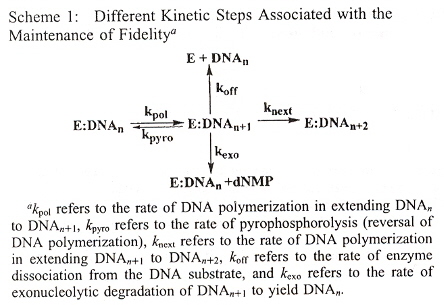
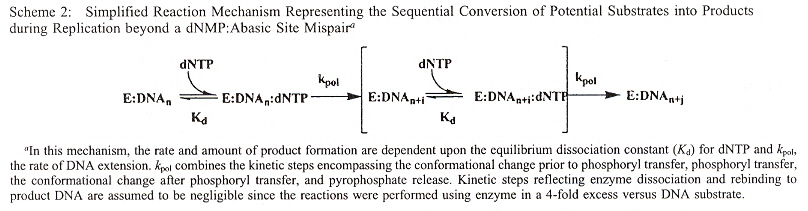
Stability of non-natural bases opposite abasic sites are examined by their effects upon the ability of polymerization to efficiently replicate, using kinetic studies (rates of reaction). Abasic lesions are used to measure replication, as potential templating information originally present in the DNA has thus been lost. The results found:
- The size of the non-natural nucleobase does not correlate with increased insertion efficiency opposite abasic sites
- hydrogen bonding is not necessary for nucleobase insertion
- base-stacking or desolvation of the incoming nucleobase is of predominant importance
- Using Rev 1 polymerase with pol ξ, dCMP insertion is favored across the abasic lesion
- Using Pol η and pol ι, dGMP insertion is favored across the abasic lesion
- With most DNA polymerases, dAMP insertion is favored across the abasic lesion (A-rule)
Steady state rate measurements were curve-fitted to the linear areas (first-order), using the formula: y = mt + b. During polymerization, non-linear burst were observed, using the equation y = Ae-kt + Bt + C where "A" is the burst amplitude, and "B" is the steady-state rate.
The observed rate is linear
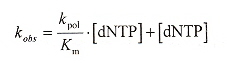
| Abbreviation | Meaning |
| dNTP | natural deoxynucleoside triphosphate |
| dXTP | nonnatural deoxynucleoside triphosphate |
| 5-NITP | 5-nitroindolyl-2'-deoxyriboside triphosphate |
| Ind-TP | indolyl-2'-deoxyriboside triphosphate |
| 5-PhITP | 5-phenyl-indolyl-2'-deoxyriboside triphosphate |
| 5-FITP | 5-fluoro-indolyl-2'-deoxyriboside triphosphate |
| 5-AITP | 5-amino-indolyl-2'-deoxyriboside triphosphate |
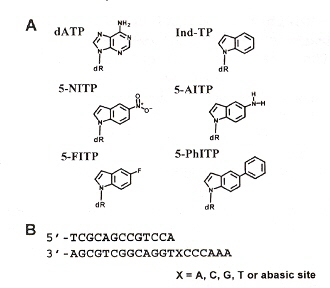
The observed kinetic rates are found in Table 1, and with a lack of significant "burst", dXTP are incorporated across the nontemplating abasic sites depending more upon π-electron surface area than shape complementarity.
Rate of Nontemplating Base Incorporation
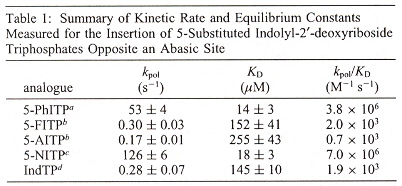
"The Use of Nonnatural Nucleotides to Probe the Contributions of Shape Complementarity and π-Electron Surface Area during DNA Polymerization", by X. Zhang, I. Lee, A. J. Berdis, Biochemistry, Oct. 4 2005, 44, 39, 13101 - 13110
"Evaluating the Contribution of Base Stacking during Translesion DNA Replication", by E. Z. Reineks, A. J. Berdis, Biochemistry, 2004, 43, 393 - 404
"Dynamics of Translesion DNA Synthesis Catalyzed by Bacteriophage T4 Exonuclease-Deficient DNA Polymerase", by A. J. Berdis, Biochemistry, June 19 2001, 40, 24, 7180 - 7191
"Simple Fluorescent Pyrimidine Analogues Detect the Presence of DNA Abasic Sites". by N. L. Greco, Y. Tor, Journal of the American Chemical Society, Aug. 10 2005, 127, 31, 10784 - 10785
Fluorescent nucleoside analogues can be used to mark the presence or absence of abasic sites. A simple fluorescent analogue of T displays different fluorescent emissions depending upon the presence or asbsence of abasic sites.
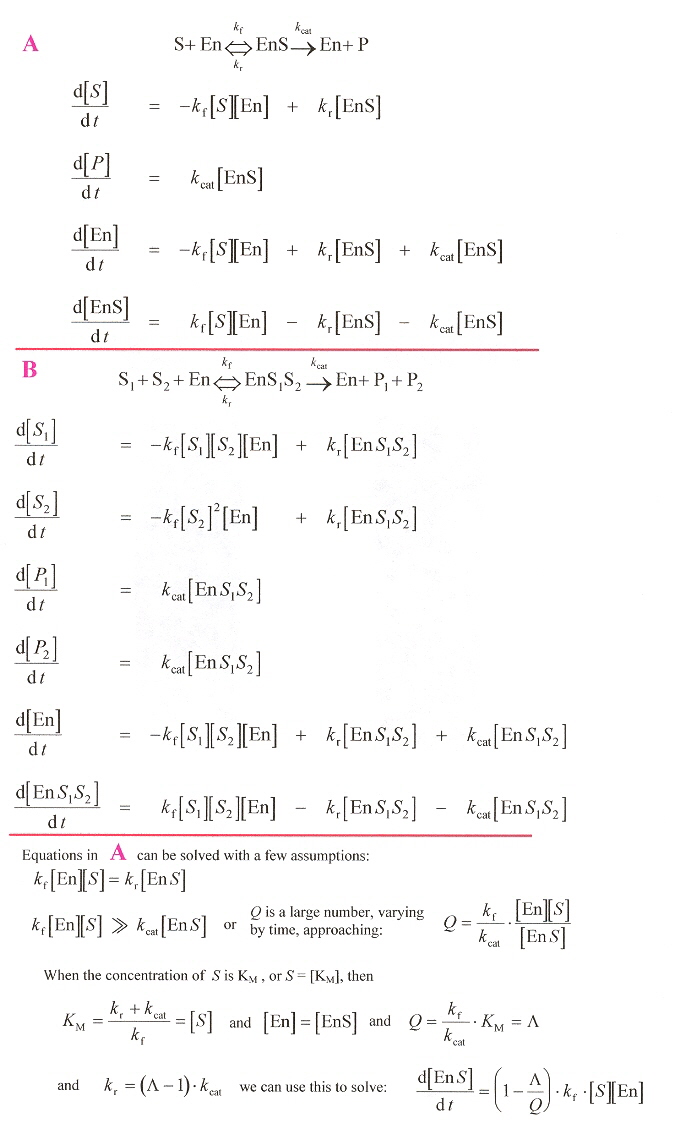
"A enzyme mechanism language for the mathematical moderling of metabolic pathways", by C.-R. Yang, B. E. Shapiro, E. D. Mjolsness, G. W. Hatfield, Bioinformatics, 2005, 21, 6, 774 - 780
This paper examines enzyme catalysis rates and mechanisms. The paper examines systems of ordinary differential equations. The only claim of covering "languages" is the use of MathematicaTM or alternatively, the use of a SBML (Systems Biology Markup Language). If SBLM is like HTML, the language capability is Chomsky type 3 (the most simple type 3, as there are only a small, finite number of "commands"). Thus there is absolutely nothing of significance in this paper from the point of view of languages (which is altogether very painfully obvious). However, the catalytic equations are of interest.
Enzyme Rates
Point to RNA, Pseudoknots, etc. Bond shifts from nitrogen at position "1" to carbon at position "5".
Non-Typical RNA Oligonucleotidesa>
"Molecular Biology of the Gene", by J. D. Watson, W. A. Benjamin, Inc., New York, 1965, p. 321
Back to TopAmino acids/Proteins
"Endonucleolytic processing of covalent protein-linked DNA double-strand breaks", by M. J. Neale, J. Pan, S. Keeney, Nature, Aug. 18 2005, 436, 7053, 1053 - 1057
DNA double-strand breaks with protein Spo11 covalently attached to 5' strand termini to initiate meiotic recombination. These double-strand breaks must be repaired by removing the Spo11 proteins. A general asymmetric process is proposed and discussed in this paper.
"Unexpected amino acid composition of modern Reptilia and its implications in molecular mechanisms of dinosaur extinction", by G-Z Wang, B-G Ma, Y. Yang, H-Y Zhang, Biochemical and Biophysical Research Communications, August 12, 2005, 33, 4, 1047 - 1049
The protein sequences for classes Ciliophora, Cnidaria, Platyhelminthes, Nematoda, Annelida, Mollusca, Arthropoda, Eichinodermata, Actinopterygii, Amphibia, Aves, Mammalia are compared to the class Reptilia. Rare or mutant or variant amino acids such as Asx or B (Aspartic acid or Asparagine), Glx or Z (Glutamine or Glutamic acid), {HCY}, {HSE}, {NLE}, {NVA}, {ORN}, {PEN}, {pGLU}, {dnp-LYS}, {pTHR}, {pSER}, {pTYR}, {gamma-GLU}, {CIT}, {nme-ALA}, {nme-ILE}, {nme-LEU}, {nme-PHE}, {nme-VAL}, {nme-SER}, {nme-THR}, {nme-TYR}, {alpha-ABA}, {iso-ASP}, {Ac-LYS}, {2-Me-ALA}, {OXA}, Cyclosporine derived MeBmt, MeBma, etc. are excludeda. A difference in distribution in CvP-bias (charged versus polar or non-charged amino acids) found in reptiles and birds is taken as temperture-adaptive and explaining the extinction of the closely related dinosaurs (without this temperature adaptation).
a Using International Union of Pure and Applied Chemistry (IUPAC) amino acid codes.
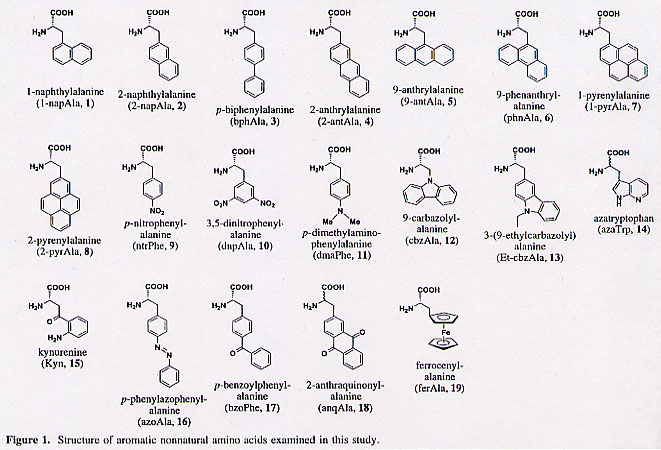
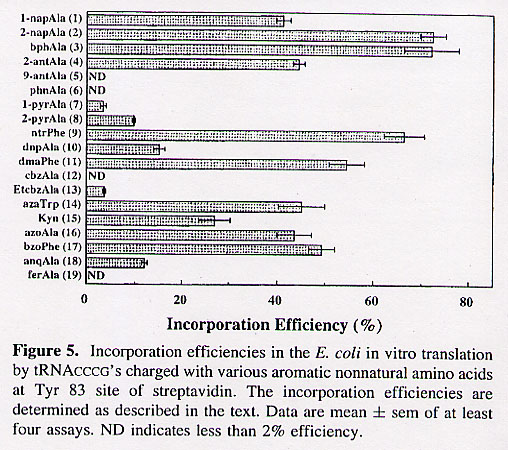
"Efficient Incorporation of Nonnatural Amino Acids with Large Aromatic Groups into Streptavidin in In Viro Protein Synthesizing Systems", by T. Hohsaka, D. Kajihara, Y. Ashizuka, H. Murakami, M. Sisido, Journal of the American Chemical Society, 1999, 121, 34 - 40
Nonnatural amino acids carrying various aromatic groups may serve as fluorescent probes. The nonnatural amino acids examined so far have been simple, restricted to small side groups. Figure 1 shows some of the nonnatural amino acids examined in this paper. In addition, Figure 5 shows how efficiently these nonnatural amino acids are expressed.
"Structure-Activity Studies of 14-Helical Antimicrobal β-Peptides: Probing the Relationship between Conformational Stability and Antimicrobal Potency", by T. L. Raguse, E. A. Porter, B. Weisblum, S. H. Gellman, Journal of the American Chemical Society, 2002, 124, 12774 - 12785
Multicellular organisms employ diverse, short peptides to defend against microbal invaders. Host-defense peptides are usually cationic, and they appear to act by permeablizing microbal membranes. Cationic charge directs the peptides to anionic bacterial membranes, and hydrophobic side chains then interact with the core of the lipid bilayer, ultimately compromising the barrier function of the membrane. Amphiphilic helix-forming oligomers of β-amino acids ("β-peptides") that display varying degrees of antimicrobal activity have recently been reported. Earlier work showed that β-peptides can adopt several distinct helical conformations, depending upon residue substitution pattern. However, peptides that disrupt bacterial membranes cannot have therapeutic utility unless they leave human cell membranes intact (generally evaluated via hemolysis). Ten interesting β-peptides are of interest here.
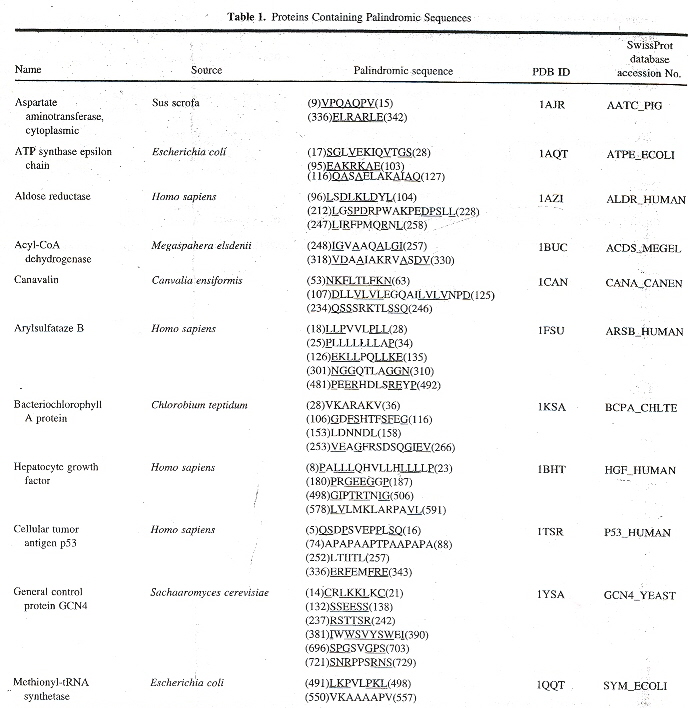
"Palindromes in Proteins", by M. Giel-Pietraszuk, M. Hoffmann, S. Dolecka, J. Rychlewski, J. Barciszewski, Journal of Protein Chemistry, Feb. 2003, 22, 2, 109 - 113
There is a slight problem with this paper: the title suggests that protein palindromes are the subject, but the authors quickly shift our search to "palindromic" sequences (meaning palindromes or almost palindromes). Furthermore, what are sought are really correctly described as palindromic sub-sequences of amino acids. The authors point out that "...Ala-Gly is not totally equivalent to Gly-Ala. It means that in the case of proteins, one can discuss palindrome-like sequence without geometric symmetry." It isn't clear that the symmetry referred to being in the linear amino acids sequence or the geometry of the corresponding 3-dimensional protein (a very significant distinction: are we seeking 2-dimensional palindromic structures or 3-dimensional palindromic structures)? In the 3-dimensional case, an analysis might require a 3-dimensional shape grammar, for example. The authors soon clear up our questions with an example.
The authors point out that the following is what is meant for AspRS
- (74)APAPAAPTPAAPAPA(88) meaning 74 amino acids, then the palindrome APAPAAPTPAAPAPA, followed by 88 amino acids
- (252)LTIITL(257) meaning 252 amino acids, followed by the LTIITL palindrome, followed by 257 amino acids
- (336)ERFE♦MFRE(343) meaning 336 amino acids, then ERFE, a number of amino acids, then MFRE, followed by 343 amino acids, where ERFE is "almost" MFRE in reverse
The authors of this interesting paper point out that palindrome sub-sequences as well as palindromic sub-sequences most likely are very significanta, by listing a number of proteins with palindromic sub-sequences found in the Brookhaven Protein Data Base (PDB). However, 2-dimensional and 3-dimensional palindromes that could appear in tertiary folding are not examined. A few examples follow. The first is a well known Roman example.
|
becomes |
|
A few other cells that can be made into 2-dimensional palindromes
TRAP RATS CARES FARADRAJA ABUT ANELE ALEDA
AJAR TUBA REFER REBER
PART STAR ELENE ADELA
SERAC DARAF
Protein Palindromic Sequences
a "Telomerase- and recombination-independent immortalization of budding yeast", by L. Maringele, D. Lydall, Genes & Development, 2004, 18, 2663 - 2675
The paper by Maringele and Lydall supports the view that palindromes are very significant. The view that short of telomeric structures and recombination, palindromes induce large DNA palindromes, and thus form as a consequence of post-senescence growth (and thus act much as telomerases do).
Continuing the discussion about 2- and 3-dimensional palindromes, this paper does not take into account such higher dimensional palindromes. Higher dimensional palindromes would appear as somewhat spacially distant. Such palindromes could appear in DNA, RNA (secondary and tertiary structures), telomeric quadruplexes, pentaplexes, hexaplexes, as well as in oligopeptides, including PNA, DNA/PNA hybrides, etc. For the moment, consider some 3-dimensional palindromes. Often, 3-dimensional palindromes have special requirements: repeated "letters". The first example below requires a string of "R". In fact, when "R" represents an amino acid, "R" need not represent a single amino acid, but any of a class of amino acids with similar properties such as approximate size, the same hydrophilic or hydrophobic properties, sulphur atoms, aryl group, etc. Similarly, when dealing with DNA or RNA, similarity such as derivatives (C vs iso-C or methylated C), etc. allow a "class" of molecules to be used, thus a string of "R" need not be so restrictive. Now for some examples of 3-dimensional palindromes. In addition to the above, consider protein knots. Such knots can change the conformation of a protein, bringing telomeric units into proximity, or removing proximal locations. Thus such knotted structure can be related to palindromic relationships as well.
Three cells (below), α, β, and γ that can be used to make 3-dimensional palindromes.
|
|
|
The second 4 × 4 cell (above) is β and will be used to construct a 3-dimensional palindrome. Imagine level 1 on top of level 2, on top of level 3, on top of level 4 (see below), then this is a 3-dimensional palindrome in Alu, Arg, Cys, Asp.
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
Note that for 3-dimensional palindromes, some repititions might be neccessary (as in cells β and γ). This can be eased up if the elements (amino acids in this case) are "palindromic" (ie: are not palindromes, but are "almost" palindromes).
"Selenocysteine tRNA identification in the model organisms Dixctyostelium discoideum and Tetrahymena thermophilia", by R. K. Shrimali, A. V. Lobanov, X-M Xu, M. Rao, B. A. Carlson, D. C. Mahadeo, C. A. Parent, V. N. Gladyshev, D. L. Hatfield, Biochemical and Biophysical Research Communications, April 1 2005, 329, 1, 147 - 151
The tRNA used to insert selenocysteine into protein in protozoans Dixctyostelium discoideum and Tetrahymena thermophilia is seryl-tRNA, designated tRNA[Ser]Sec, and the insertion takes place when the UGA codeword is detected. Comparison of selenocysteine tRNA in different organisms shows a distinct lack of homology, indicating that this molecule has undergone substntial evolutionary change.
"Pyrrolysine and Selenocysteine Use Dissimilar Decoding Strategies", by Y. Zhang, P. V. Baranov, J. F. Atkins, V. N. Gladyshev, The Journal of Biological Chemistry, May 27 2005, 280, 21, 20740 - 20751
A study that focuses upon:
- Selenocysteine (Sec), the 21st amino acid, which reads through what is usually the UGA stop codon, using the SECIS insertion sequence, and
- Pyrrollysine (Pyl), the 22nd amino acid, which reads through what is usually the UAG stop codon, using the PYLIS insertion sequence
Note that selenocysteine is often found in Methanogenic archaea.
"Selenoproteins—Tracing the Role of a Trace Element in Protein Function", by T. C. Stadtman, Public Library of Science Biology (PLOS Biology), Dec. 2005, 3, 12, (E421) 2077 - 2079
In mammals, methionine-R-sulfoxide reductase (MsrB1) contains selenocysteine at the active site, while MsrB2 and MsrB3 contain cysteine at the active site. MsrB2 and MsrB3 convert cysteine thiol to sulfenic acid (SOH) which in turn is reduced by thioredoxin to regenerate cysteine. However, MsrB1 converts ionized selenocysteine to selenic acid (SeOH). SeOH is converted to selenosulfide which with thioredoxin then regenerates SeOH.
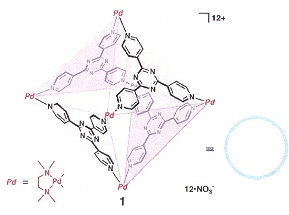
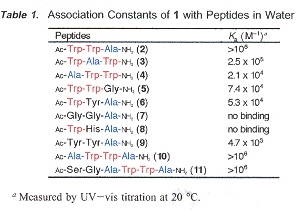
"Sequence-Selective Recognition of Peptides within the Single Binding Pocket of a Self-Assembled Coordination Cage", by S. Tashiro, M. Tominaga, M. Kawano, B. Therrien, T. Ozeki, M. Fujita, Journal of the American Chemical Society, April 6 2005, 127, 13, 4546 - 4547
Sequence-selective recognition of peptides, apable of differentiation between oligopeptides is essential to the nderstanding of protein function. This paper is an exploration of this recognition problem within caged molecular structures with a pocket.
|
Cage-Molecule
|
Differential-Binding of Oligopeptides
|
Adducts
"Kinetics of Nucleotide Incorporation Opposite Polycyclic Aromatic Hydrocarbon-DNA Adducts by Processive Bacteriophage T7 DNA Polynerase", by H. Zang, T. M. Harris, F. P. Guengerich, Chemical Research in Toxicology, Feb. 2005, 18, 2, 389 - 400
Polycyclic aromatic hydrocarbons (PAHs) are widespread envionmental contaminants found in tobacco smoke, automobile exhausts, food, etc. that cause mutation, such as to tumor suppressor gene p53. This paper investigates the possibility of bypassing dA and dG PAH adducts during DNA replication, using bacteriophage T7 DNA polymerase and reverse transcription. The results found are that bulky PAH adducts are highly disruptive to DNA replication.
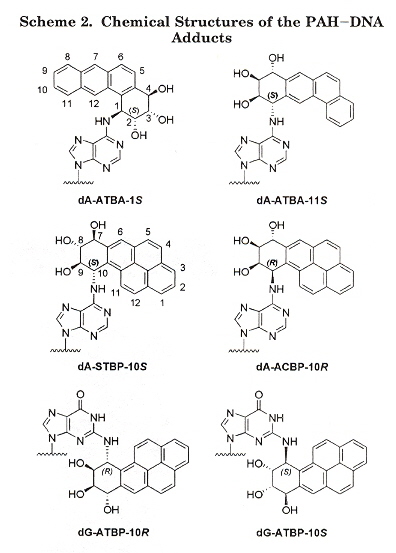
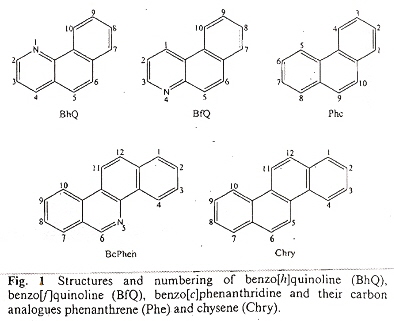
"Theoretical study of aza-polycyclic aromatic hydrocarbons (aza-PAHs), modelling carbocations from oxidized metabolites and their covalent adducts with representative nucleophiles", by G. L. Borosky, K. K. Laali, Organic and Biomolecular Chemistry, 2005, 3, 7, 1180 - 1188
In addition to PAH adducts, aza-PAH adducts (PAHs with nitrogen incorporated into aromatic rings) are mutagenic and carcinogenic. A few examples of aza-PAHs follows.
Examples of aza-PAH Adducts
DNA Damage Repair
"Spontaneous Homologous Recombination Is Induced by Collapsed Replication Forks That Are Caused by Endogenous DNA Single-Strand Breaks", by N. Saleh-Gohari, H. E. Bryant, N. Schultz, K. M. Parker, T. N. Cassel, T. Helleday, Molecular and Cellular Biology, Aug. 2005, 25, 16, 7158 - 7169
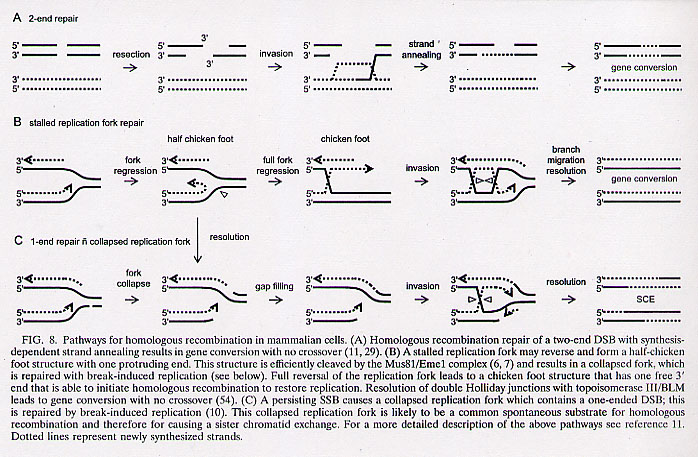
DNA damage repair during DNA replication in mammalian cells is investigated. Double-strand-breaks (DSB) resemble those of collapsed replication forks. Single-strand-breaks (SSB) are converted to double-strand-breaks in the presence of replication. Fig. 8 (below) may be used as a foundation to explain results found in this paper.
Fig. 8A explains (diagramatically) two-end repair where there are DSB.
Fig. 8B explains (diagramatically) how stalled replication fork repair can take place.
Fig. 8C explains (diagramatically) how a replication fork with a SSB takes place.
"Translocation of Nicked but not Gapped DNA by the Packaging Motor of Bacteriophage phi29", by W-D. Moll, P. Guo, Journal of Molecular Biology, August 5, 2005, 351, 1, 100 - 107
The biomolecular mechanism used by double-stranded DNA viruses to insert and package their DNA into a preformed procapsida is unknown. This mechanism was examined by structurally modifying phi29 using DNASE I to cause nicks at random sites, or using N.BbvC IA at specific sites. Site specific nicks could then be expanded using T4 DNA polymerase. What then might the differences be between nicked phi29 vs. gapped phi29? Nicked phi29 DNA was packaged as efficiently as unnicked phi29 (but the nicks were not repaired). However, gapped phi29 DNA was packaged only until a gap was encountered. Nicks can be tolerated, but gaps cannot be tolerated.
aA "procapsid" is a protein shell lacking a virus genome.
"Binding of MutS mismatch repair protein to DNA containing UV photoproducts, 'mismatched' opposite Watson-Crick and novel nucleotides, in different DNA sequence contexts", by P. D. Hoffman, H. Wang, C. W. Lawrence, S. Iwai, F. Hanaoka, J. B. Hays, DNA Repair, Aug. 8 2005, 4, 9, 983 - 993
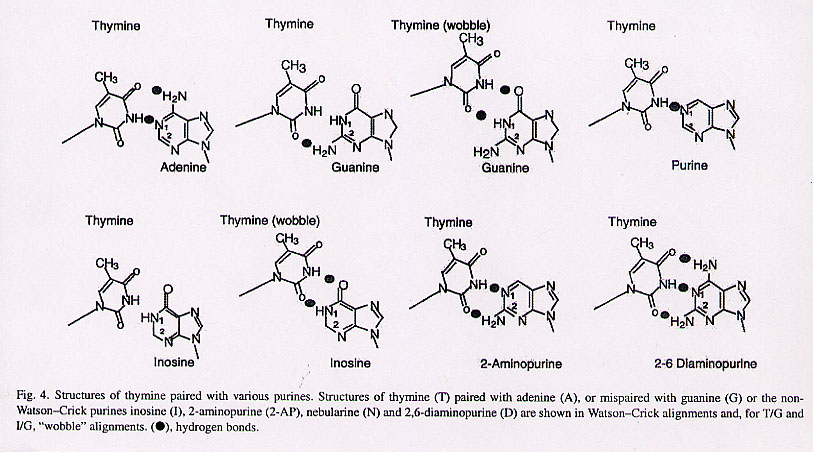
Mismatched pairs have long been discussed and are a well known theme. In this paper, of immediate relevance are discussions of some novel nucleotide mismatches. Thymine/Inosine and Thymine/Inosine wobble are augmented with T/G mismatches, T/G wobble mismatches, T/Purine mismatches, T/2-Aminopurine mismatch, and T/2-6 Diaminopurine mismatches.
Back to TopSynthetic Biology, Bioinformatics
"DNA, diseases and databases: disastrously deficient", by G. P. Patrinos, A. J. Brookes, Trends in Genetics, June 2005, 21, 6, 333 - 338
"Emergent Computation: Emphasizing Bioinformatics" is concerned with the linguistic structures in Bioinformatics (DNA, RNA, and Proteins), and does not wish to examine databases of substrings of DNA, RNA, and proteins. There already are a plethora of books dedicated to databases composed of substrings of genetic bases. It would be of interest, however, to examine briefly the limited database approach to Bioinformatics. This paper is a critical review of genetic databases.
Four types of mutation databases:
- central databases (CDBs)
- locus-specific databases (LSDBs)
- DNA polymorphisms (DNADBs)
- national mutation databases (NMDBs)
CDBs tend to include only mutations of large effect that can be described in Mendelian terms (sequence variations associated with minor clinical consequences are rarely considered). Thus CDBs correlate DNA polymorphism to phenotype.
LSDBs contain information about one or a few specific genes usually related to a single disease. Thus LSDBs correlate DNA polymorphism to phenotype.
DNADB do not correlate DNA data to phenotypes.
Major problem with all these databases is that they don't necessarily record the same data, thus may not be compatible, and are often created by needy users, thus their designs are not limited by any standards. Another problem is that they tend to be limited to a four-letter code. Yet another problem is that they exclude RNA, proteins, metabolic classes, and epigenetics. Another problem is that they are oriented more towards the medical field, and thus exclude such areas as nanotechnology applications.
"Scientists mixing, matching basic DNA building blocks", P. Elias, San Jose Mercury News, August 22, 2005, 3E ("Life Engineering Symposium").
Large scale mixing of genes and DNA from various organisms called "synthetic biology" to create or engineer new viruses, or organisms capable of producing alternative fuels, malarial drugs, insulin, anti-cancer therapies, molecular sensors, DNA motors, RNA switches, synthetic bacteriological delivery agents, etc.
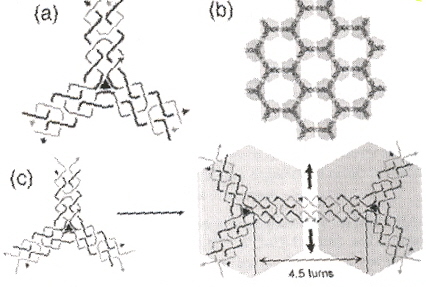
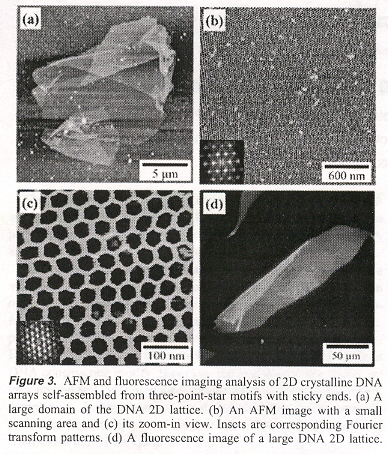
"Self-Assembly of Hexagonal DNA Two-Dimensional (2D) Arrays", by Y. He, Y. Chen, H. Liu, A. E. Ribbe, C. Mao, Journal of the American Chemical Society, Sept. 7 2005, 127, 35, 12202 - 12203
A three-pointed-star DNA motif that can self-assemble into porous, hexagonal, two-dimensional (2D) arrays is reported. If rotated 90°, essentially, the same results were predicted using a Shape grammar in "Emergent Computation: Emphasizing Bioinformatics" in the Appendix, p. 387.
|
Hexagonal: Shape grammar
|
Experimental
|
"The Language of Life: How Cells Communicate in Health and Disease", by D. Niehoff, Nature, June 30 2005, 435, 7046, 1162
In this book review by F. Balkwill, the notion that language can be applied to cellular societies and in the area of molecular studies is discussed. Language based not upon sounds or gestures, but rather replacing words and sentences with proteins that have aspects similar to human psychological functions. Apparently a popular approach to studying language in nature that does not transcend the view that such languages are more than descriptive, that such languages for cellular societies and molecules may actually be factually inherrent, and that a psychological component of language is not necessary.
Back to Top