"Ribosome Gymnastics - Degree of Difficulty 9.5, Style 10.0", by J. F. Atkins, R. B. Weiss, R. F. Gesteland, Cell, Aug. 10 1990, 62, 413 - 423
Ribosomes appear to have three sites: A, P, and E (exit). Ribosomal functions include:
- Slipping (frameshifting)
- Hopping (frameshifting)
- Stop codon Readthrough
Typically, triples of bases (codons) are read. Sometimes, paired tRNA anti-codon: mRNA codon disengage, allowing the mRNA to "slip". The tRNA anticodon may then re-pair with a frameshifted mRNA codon. Most often, these "slippages" are small (+1 base, or -1 base), but slippages of many bases can take place. An example of a "slippery" run (four or more repeated bases): A AAA AAC
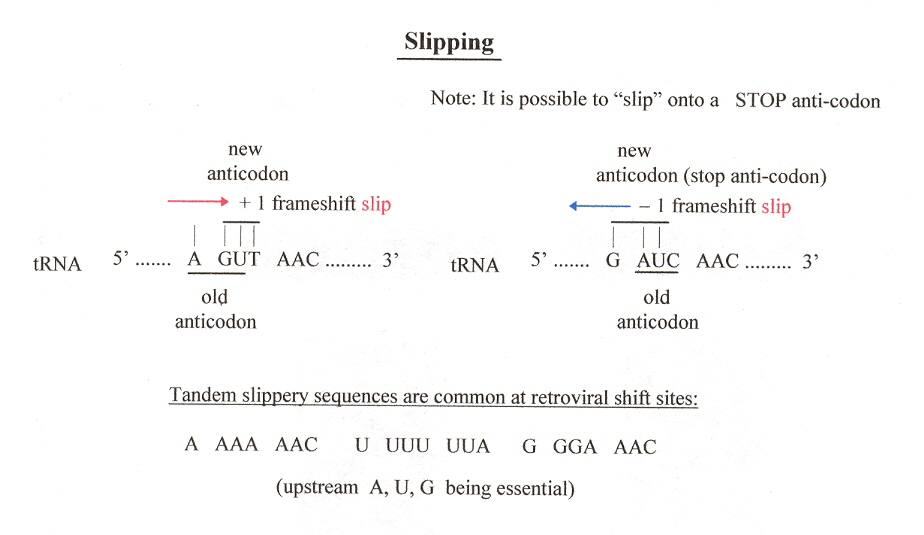
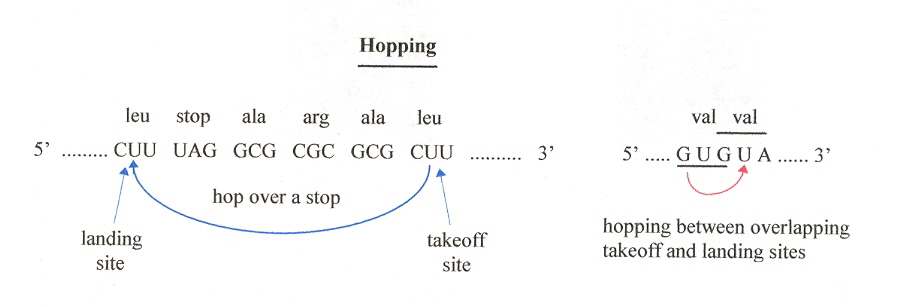
If slipping occurs over many bases without re-pairing of tRNA anticodon:mRNA codon, then this is referred to as "hopping". Hopping typically has a "takeoff codon site" and a "landing codon site". Example:
"Exon skipping and reading through stop codons: two research approaches for a therapy of Duchenne's muscular dystrophy, Monaco, 17 - 18, January, 2004", The Journal of Gene Medicine, 2005, 7, 249 - 255
A very moving technical paper. Muscular dystrophy is caused by a point mutation such as the omission of a nucleotide base in exons of the Duchenne gene. The omission of a base changes the frame (triples of bases), thus causing Duchenne's muscular dystrophy. Various researchers trying to create stable anti-sense cDNA in mRNA which is capable of restoring at least partial correction in 90% of Duchenne's muscular dystrophy by cancelling out the effect of frame error(s) induced by point mutation(s) in exons. It has worked in mice, corrections being observed in less than 24 hours, maybe it will such approaches will be able to help human beings. Such corrective measures might work in many diseases caused by genetic errors. This is an example of real intelligent design.
Approaches to correct genetic frame errors include exon skipping which changes a Duchenne mutation into Becker's mutation with a less severe form of muscular dystrophy. Yet another approach to these diseases is the use of gentamicin. Mutations to the dystrophin gene change an amino acid codon into a stop codon (UGA, UAG, UAA), thus protein synthesis stops prematurely, causing Duchenne's dystrophy. Gentamicin can read-through premature stop codons, thus alleviating disease.
"DNA Drug Design for Cancer Therapy", by Y. S. Cho–Chung, Current Pharmaceutical Design, 2005, 11, 22, 2811 - 2823
Antisense synthetic Oligodeoxynucleotides that are Watson–Crick complimentary to single-stranded DNA hybridize with corresponding mRNA thereby inhibiting translation of genetic elements into proteins. DNA-based therapeutics can be accomplished with antisense methodology or the use of synthetic triple-helix decoy methods mentioned in "Emergent Computation: Emphasizing Bioinformatics" (p. 307).
Back to TopG–Quartets in Medicine
"Rational Drug Design of G–Quartet DNA as Anti–Cancer Agents", by N. Jing, W. Sha, Y. Xiong, D. J. Tweardy, Current Pharmaceutical Design, 2005, 11, 22, 2841 - 2854
A new class of non-antisense DNA agents based upon formation of G–quartet structures in G-rich DNA. The quartet structures interfere with target proteins. Used as anti-cancer agents, since telomeric DNA contains (TTAGGG)n capable of forming G-quartets, and such quartets were shown to be telomerase inhibitors in tumor cells in vitro.
Back to Top