DNA/RNA & α-Amino acid Peptide Mimetics
"Sugar amino acids in designing new molecules", by T. K. Chakraborty, P. Srinivasu, S. Tapadar, B. K. Mohan, Glycoconjugate Journal, March 2005, 22, 3, 83 - 93
A number of papers have been published which focus upon the construction of DNA or RNA mimetic molecules. Such molecules might have a variety of uses, such as studying alternate molecular bases of life other than DNA or RNA (such as the TNA discussed in "Emergent Computation: Emphasizing Bioinformatics"), or pharmaceuticals in which such mimetics might form double strands with DNA or RNA or even triple strands. In this paper, sugar amino acids (Saa) are studied. The authors cite similar work done by other researchers, and it is convenient to cite some examples of such other research here, as well.
"Vinylogous Polypeptides: An Alternative Peptide Backbone", by M. Hagihara, N. J. Anthony, T. J. Stout, J. Clardy, S. L. Schreiber, Journal of the American Chemical Society, 1992, 114, 6568 - 6570
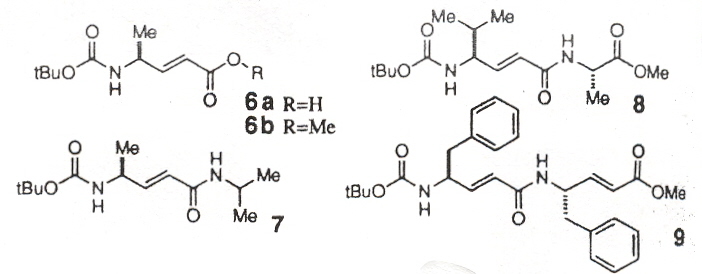
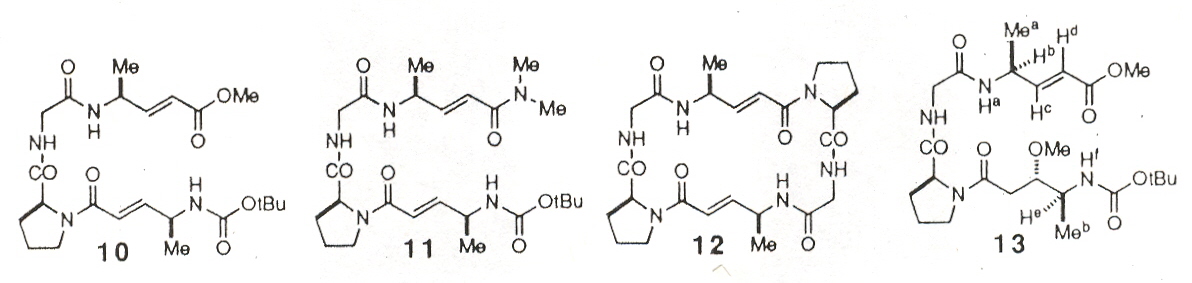
This paper examines the secondary and tertiary structure of polypeptide chains based not upon amino acids, but upon derivatives of amino acids. In particular, amino acids with an ethenyl unit between the carbonyl carbon and the alpha carbon (vinylogous amino acids). Molecule 6b was found to pack without a highly ordered secondary structure, but changing the oxygen ester to the NH amides of molecules 7, 8, 9 resulted in long stacks of parallel sheets. Similarly, the change from the oxygen ester to the amide in molecule 10 vs molecule 11. Molecule 11 likely forms antiparallel sheets. Molecule 13 adopts a helical conformation similar to that found in normal peptides.
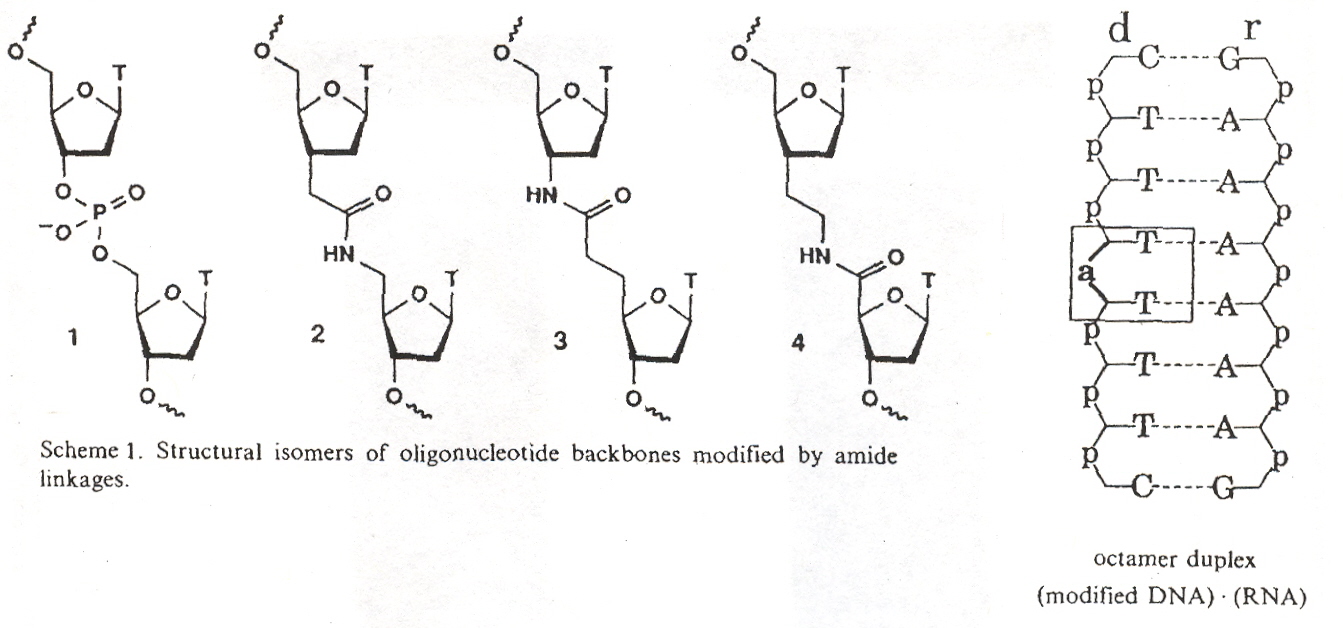
"Amides as a New Type of Backbone Modification in Oligonucleotides", by A. D. Mesmaeker, A. Waldner, J. Lebreton, P. Hoffmann, V. Fritsch, R. M. Wolf, S. M. Frier, Angewandte Chemie International Edition", 1994, 33, 2, 226 - 229
Oligonucleotides containing phosphoramidites (mixed amide/phosphor backbones) were synthesized. DNA:RNA heteroduplexes containing amide linkages were synthsized.
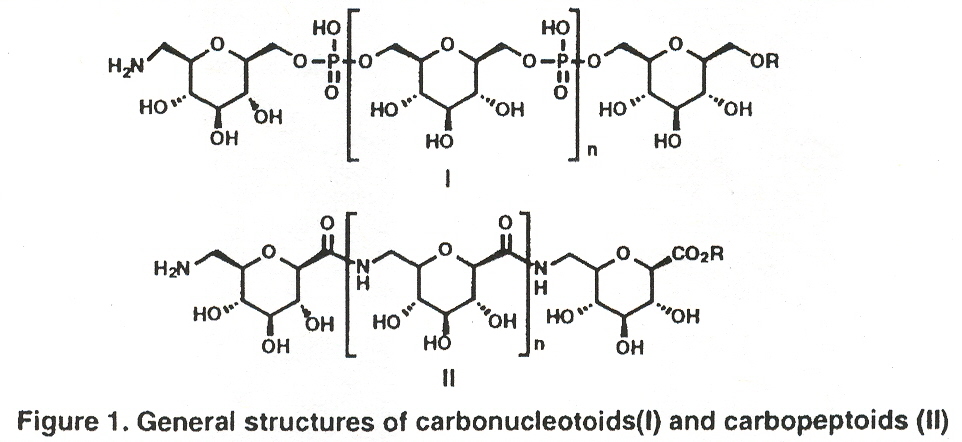
"Carbonucloetides and Carbopeptides: New Carbohydrate Oliomers", by K. C. Nicolau, H. Flörke, M. G. Egan, T. Barth, V. A. Estevez, Tetrahedron Letters, 1995, 36, 11, 1775 - 1778
Carbopeptoids (peptide-bond linked carbohydrates) as well as carbonucleotoids (phosphate-bond linked carbohydrates) have the potential of forming macromolecular backbones.
"Synthesis of Sulfated β-1,6-Linked Oligosaccharide Mimetics: A Novel Potent Inhibitor of HIV Replication", by Y. Suhara, M. Ichikawa, J. E. K. Hildreth, Y. Ichikawa, Tetrahedron Letters, 1996, 37, 15, 2549 - 2552
This paper discuses possible pharmaceutical applications of a class of carbopeptoids.
"The Fourth Helical Secondary Structure of β-Peptides: The (P)-28-Helix of a β-Hexapeptide Consisting of (2R,3S)-3-Amino-2-hydroxy Acid Residues", by K. Gademann, A. Häne, M. Rueping, B. Jaun, D. Seebach, Angewendt Chemie International Edition, 2003, 42, 13, 1534 - 1537
Fourth type of helical structure reported for β-peptides. These molecules are expected to have protein-like functions.
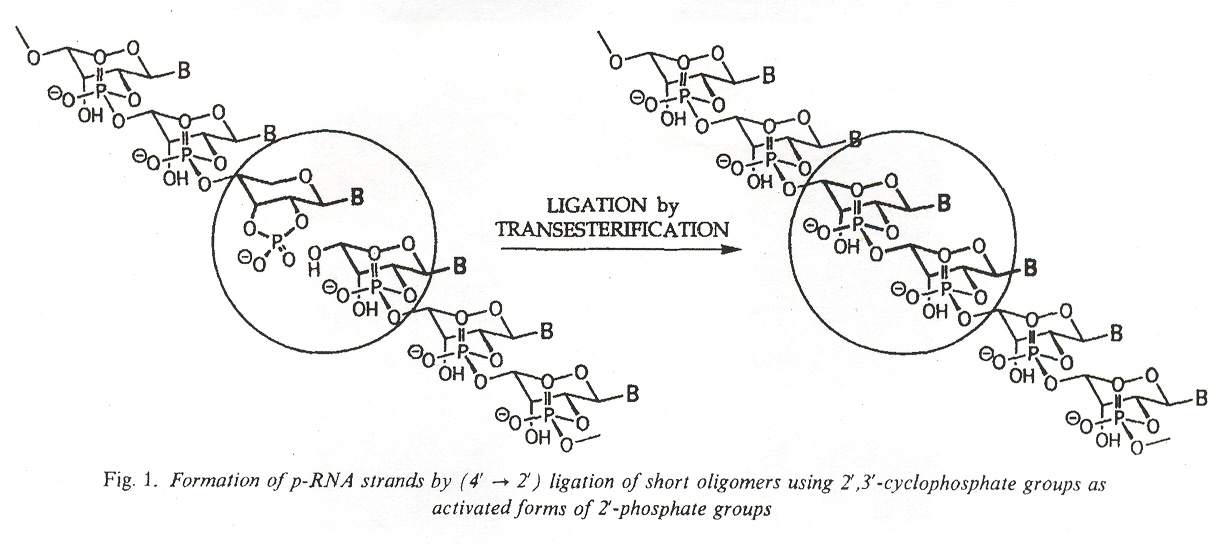
"Pyranosyl-RNA: Further Observations on Replication", by M. Bolli, R. Micura, S. Pitsch, A. Eschenmoser, Helvitica Chimica Acta, 1997, 80, 1901 - 1951
Pyranosyl-RNA (p-RNA) oligomers with bases in the form of dimers, trimers, tetramers, pentamers, hexamers, octamers, and dodecamers have been synthesized. In many cases, the molecules have been restricted to specific sequences and their complements, such as pr(GGGCGGGC) and pr(CCCGCCCG). Also AAATAAAT and TTTATTTA and other sequences were studied. p-RNA duplexes such as pr(AAAAUUUU) and pr(UUUUAAAA) were also studied. Stacking studies were also made. Yields examined. Mismatching bases studied.
"Theoretical and Experimental Circular Dichroic Spectra of the Novel Helical Foldamer Poly[(1R,2R)-trans-2-aminocyclopentanecarboxylic acid]", by J. Applequist, K. A. Bode, D. H. Appella, L. A. Christianson, S. H. Gellman, Journal of the American Chemical Society, 1998, 1 20, 4891 - 4892
The folding patterns of short oligomers of β-amino acids (β-peptides) called "foldamers" are examined. Specifically, (1R,2R)-trans-2-aminocyclopentanecarboxylic acid is examined. Results found for these β-amino acids are helical or sheet secondary-structures. Foldamers are good candidates for three dimensional shape grammars.
Back to TopPNA: Peptide Nucleic Acids
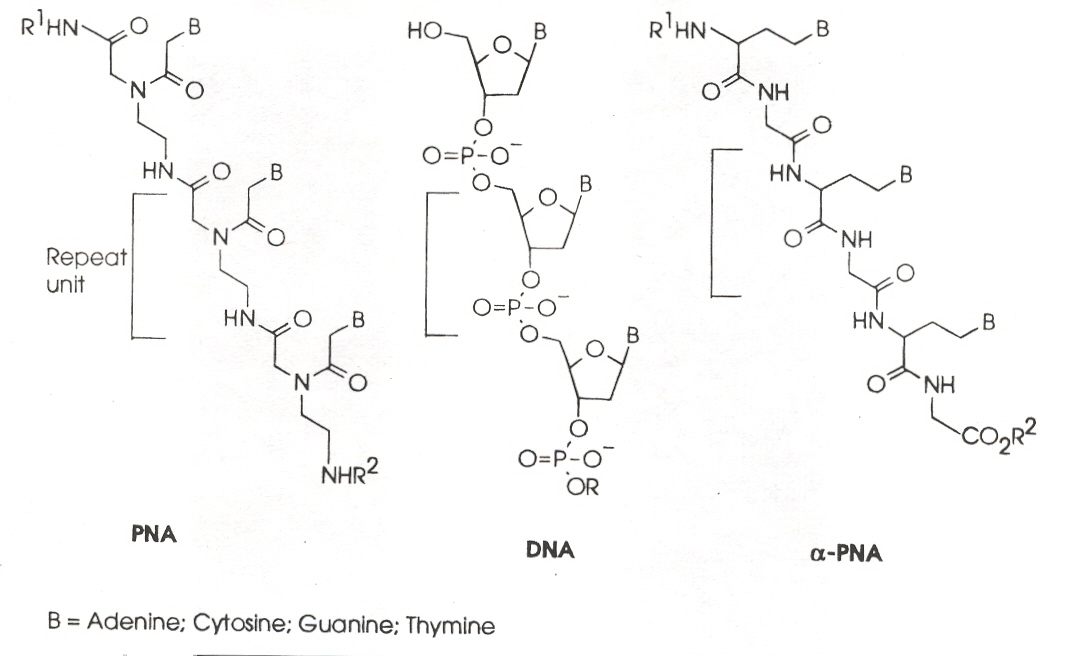
"α-PNA: A Novel Peptide Nucleic Acid Analogue of DNA", by N. M. Howarth, L. P. G. Wakelin, Journal of Organic Chemistry, 1997, 62, 5441 - 5450
PNA (peptide nucleic acid) analogues of DNA might form pharmacological regulators of gene expression as they have the capacity to form PNA:DNA heteroduplexes [and in some cases, (PNA)2:DNA triplexes]. However, PNA:DNA heteroduplexes are limited to homopurine and homopyrimidine sequences. This paper reports research synthesizing α-PNA which incorporate all four bases (A, C, G, T).
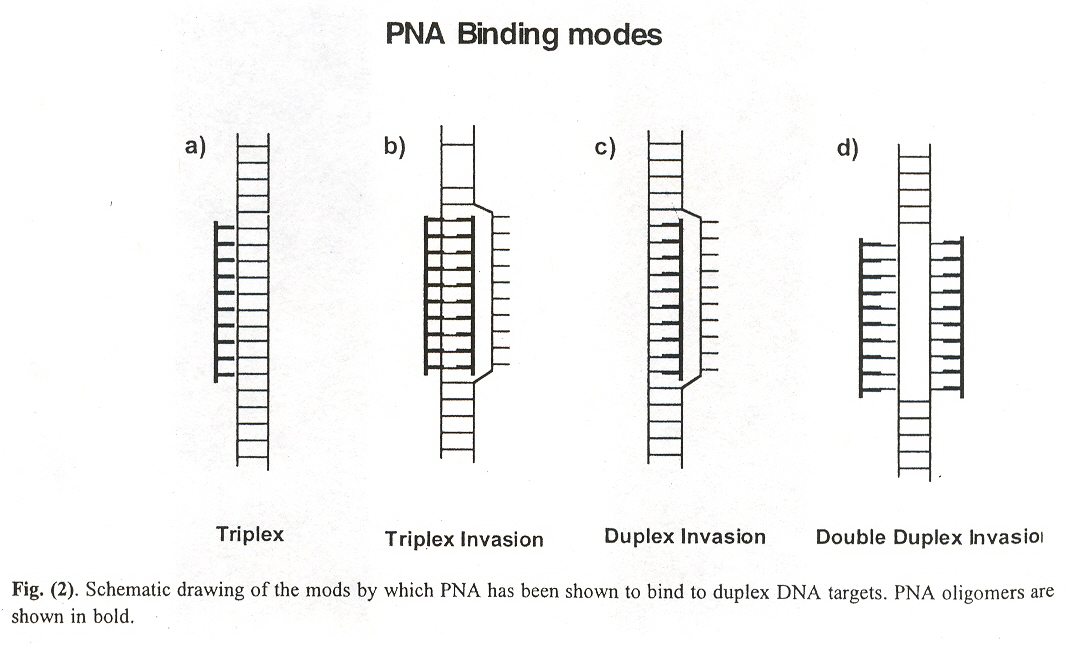
"Peptide Nucleic Acids: Analogs and Derivatives", by K. N. Ganesh, P. E. Nielsen, Current Organic Chemistry, 2000, 4, 931 - 943
Peptide Nucleic Acids (PNA) are an extremly efficient structural mimic of DNA and RNAa, but do not bind efficiently to double stranded DNA by triplex formation, but rather prefer an alternative structure, triplex invasion, see below. In fact, PNA:DNA and PNA:RNA duplexes are more stable than DNA:DNA or DNA:RNA duplexes. Triplex invasion is yet another mechanism that goes well beyond the mechanisms covered by Splicing Systems, and Dominoes and provides further evidence of the weakness of mathematical methods with a bias against biochemical fact in favour of "elegance". Note that some DNA repair mechanisms entail triplex invasion b.
aPNAs have been proposed as possible pre-RNA prebiotic systems, and PNAs have been in meteorites. "Peptide nucleic acids rather than RNA may have been the first genetic molecule", K. E. Nelson, M. Levy, S. L. Miller, Proceedings of the National Academy of Sciences of the USA, Apr. 11 2000, 97, 8, 3868 - 3871
b "Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion", by M. McVey, J. R. LaRocque, M. D. Adams, J. J. Sekelsky, Proceedings of the National Academy of Sciences of the USA", Nov. 2 2004, 101, 44, 15694 - 15699
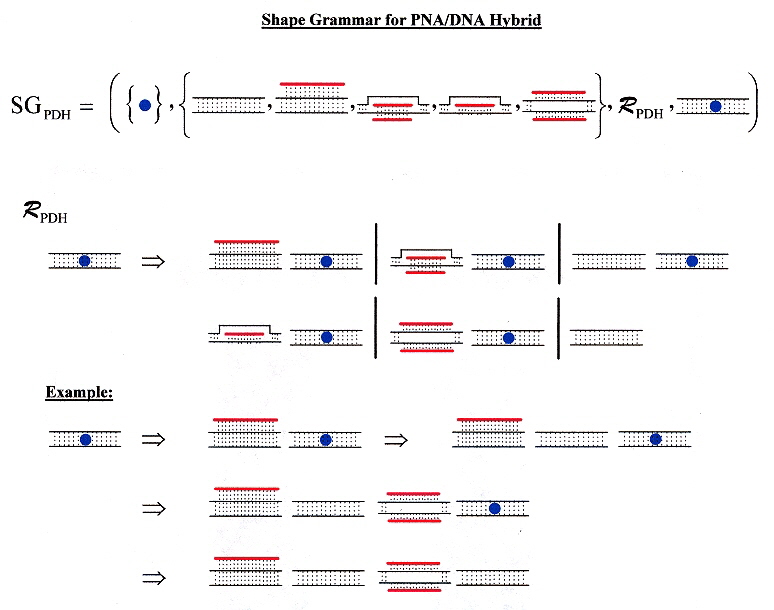
A sample shape grammar for PNA/DNA hybrids. The heavy strand is the strand of PNA with nucleotide bases.
PNA/DNA hybrid shape grammar
"End invasion of peptide nucleic acids (PNAs) with mixed-base composition into linear DNA duplexes", by I. V. Smolina, V. V. Demidov, V. A. Soldatenkov, S. G. Chasovskikh, M. D. Frank-Kamenetskii. Nucleic Acids Research, 2005, 33, 17, e146 (9 pages)
The binding of PNA oligomers inside dsDNA is limited to homopurine and homopyrimidine sequences, but in this paper, it is shown that mixed-base PNA will also bind if this is limited to the termini of dsDNA. It is hoped that these findings will be of use in DNA diagnostics as well as biotechnology.
Terminal mixed-base dsDNA PNA Invasion
"PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules", by M. Egholm, O. Buchardt, L. Christensen, C. Behrens, S. M. Freier, D. A. Driver, R. H. Berg, S. K. Kim, B. Norden, P. E. Nielsen, Nature, Oct. 7 1993, 365, 6446, 566 - 568
This paper reduces the restrictions placed upon PNA/DNA hybridization very much. Experimental
evidence shows that a PNA strand can hybridize with a DNA strand using normal Watson-Crick complements,
provided that no two purines or pyrimidines are juxtaposed. Thus pairs of A/G cannot be juxtaposed
with pairs of U/T/C. Two experimental oligonucleotides are tested:
"Structural Preorganization of Peptide Nucleic Acids: Chiral Cationic Analogues with Five- or Six-Membered Ring Structures", by V. A. Kumar, European Journal of Organic Chemistry, 2002, 2021 - 2032
This paper reviews many experiments dealing with PNAs, such as PNAs with five-membered Nitrogen heterocycles, Aminoethylprolyl PNA, Pyrrolidinone PNA, etc. Peptide Ribonucleic Acids (PRNA), PNA with six-membered ring structures, etc. It is pointed out that mixed purine/pyrimidine PNA oligomers bind to both parallel and antiparallel DNA with almost equal ability. Specifically, the C-terminus of PNA is conventionally taken to be analogous to the 3' end of DNA/RNA. This ambiguity in parllel/antiparallel DNA/RNA binding modes of PNA oligomers is probably due to compatability of achiral PNA with chiral DNA/RNA.
Back to Top