A disclaimer: finding scientific articles dealing with the subjects covered in "Emergent Computation: Emphasizing Bioinformatics" requires a great deal of work and time. The major reason for this is that the subjects of Bioinformatics from the point of view of Mathematical Linguistics as well as applications of Mathematical Linguistics in areas such as Biology, Meteorology, Oceanography, Geology, Chemistry, etc. are not yet recognized as a discipline of study. As a consequence, it is not claimed that the scientific articles or books cited here constitute all the relevant articles that may have been published, just those that have come to light.
Zinc Fingers (ZFN) (or zinc finger proteins ZFP) may be used as chimeric nucleases, used to cleave DNA. Zinc Finger Methylases (ZF–MTase) are intended to be used to methylate nucleotide bases to silence gene expression.
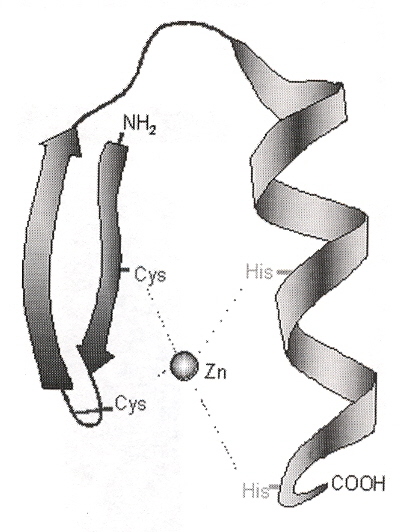
A zinc finger is a protein containing a Zinc++ cation chelated to two cysteine amino acids each on an antiparallel β sheet, and two histidine amino acids located on a single α helix (this is the ββα architecture). Such zinc fingers are of the C2H2 class. The number of amino acids between the cysteines and the histidines can be varied to form the finger of the zinc finger (to obtain conformational space). Zinc tends to create a planar coordination complex. Zinc fingers can bind to DNA (or RNA). Other less frequently encountered zinc fingers have different architectures, including C3H, C4, C6, and the artificial H2H2. Also, special studies have examined "zinc fingers" using Co2+, Ni2+ and Ce4+. While most zinc fingers have 3 fingers, some have 2 fingers, but some zinc finger proteins have up to 37 zinc fingers a. Some other zinc fingers include Zif268 (3 fingers: mice), GLI (5 fingers: humans), TTK (2 fingers: Drosophila) b. To better appreciate what is understood about zinc fingers, a numbering scheme is used to describe each finger. Specifically, the first amino acid in the α helix is designated 1, the next amino acid is designated 2, etc. until amino acid 9, the amino acid just before the α helix is designated as position –1. As an example, consider the following for Zif268 (first finger) c.
–1 1 2 3 4 5 6 7 8 9
M A E E R P Y A C P V E S C D R R F S R
S D E L T R H I R I H T
β
β
α
It is expected that zinc finger protein will have many applications in medicine. Some applications include the following d:
Specific applications have included the following e.
a "Designer Zinc Finger Proteins: Tools for Creating Artificial DNA-Binding Functional Proteins", by M. Dhanasekaran, S. Negi, Y. Sugiura, Accounts of Chemical Research, 2006, 39, 1, 45 - 52
b "Zinc fingers", by A. Klug, J. W. R. Schwabe, Federation of American Societies for Experimental Biology (FASB), May 1995, 9, 8, 597 - 604
c "Toward a code for the interactions of zinc fingers with DNA: Selection of randomized fingers displayed on phage", by Y. Choo, A. Klug, Proceedings of the National Academy of Sciences U.S.A., Nov. 23 1994, 91, 23 11163 - 11167
d "DRUG DISCOVERY WITH ENGINEERED ZINC-FINGER PROTEINS", by A. C. Jamieson, J. C. Miller, C. O. Pabo, Nature Reviews Drug Discovery, May 2003, 2, 5, 361 - 368
e "Towards therapeutic applications of engineered zinc finger proteins", by A. Klug, Federation of European Biochemical Societies Letters (FEBS Letters), Feb. 7 2005, 579, 4, 892 - 894

Citation and Abstract
"Chimeric Restriction Enzymes: What Is Next?", by S. Chandrasegarian, J. Smith, Biological Chemistry, 1999, 380, 841 - 848
DNA-binding motifs such as the zinc finger motif can be converted into a novel site-specific endonuclease by fusing the binder to the FokI cleavage domain. Zinc finger proteins bind to DNA by inserting an α-helix into the double helix. Each finger interacts with a base pair triplet. In theory, one can design a zinc finger for each of the 64 possible triplet codons. Thus one could design 'artificial' nucleases that will cut DNA at any preferred site by making fusions of zinc finger proteins to the cleavage domain of FokI endonuclease.
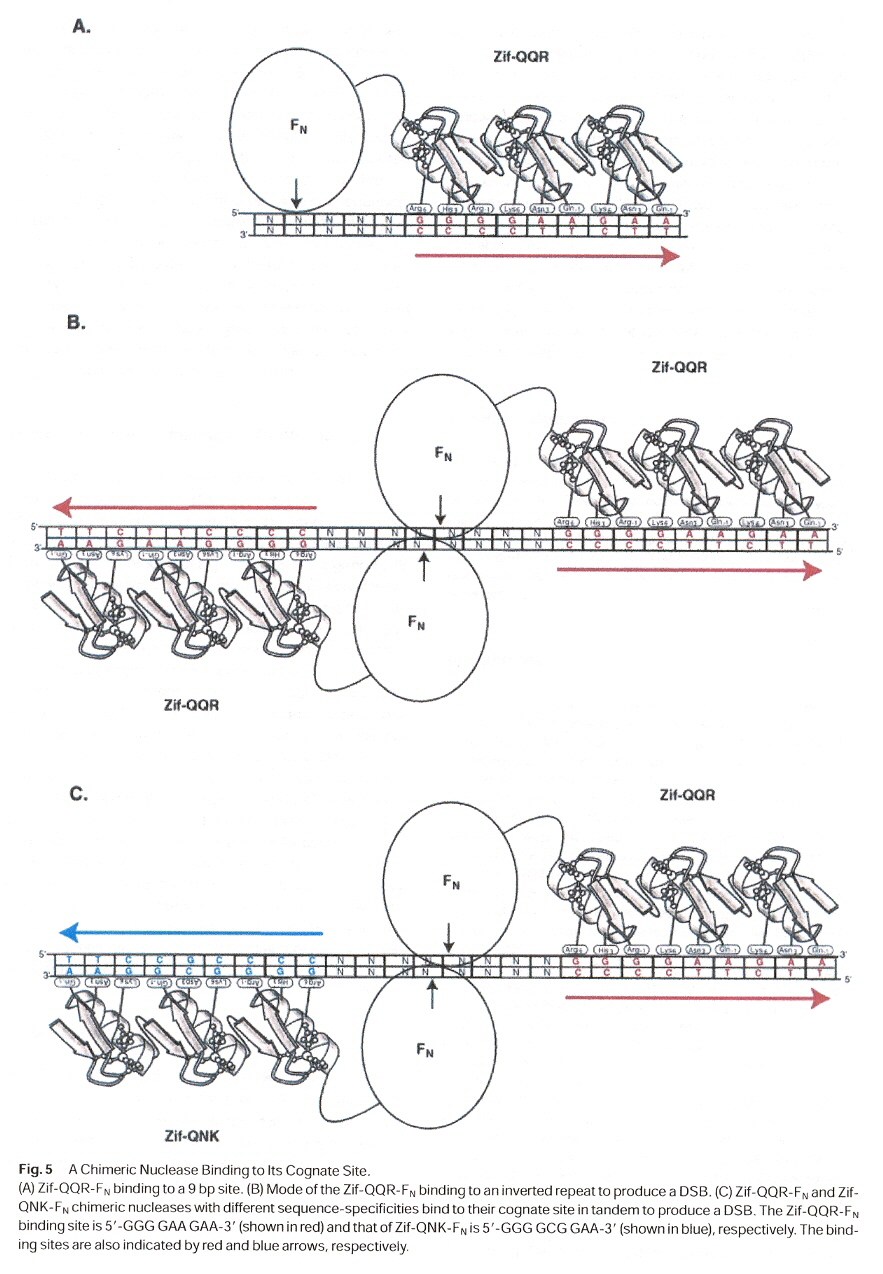
Zinc fingers may be used to create artificial endonucleases or artificial methyltransferases. One might immediately wonder if symmetry is then required? However, symmetry may not always exist in natural endonucleases or methylases. Two examples should suffice: a
Artificial zinc fingers could conceivably be constructed to cleave DNA and DNA/PNA hybrids. Not only for double-stranded DNA, or RNA, but triple-stranded as well as quadruplexes and other structures. With ligation, a world of possibilities is opened up! Artificial endonucleases that could cleave DNA triplexes, or double-stranded DNA/RNA hybrids (possibly with artificial bases too), etc. (along with ligation) completely changes such topics as Splicing Systems, Dominoes, medical and pharmacological applications, etc. Similarly, a new world of possibilities opens up for artificial methyltransferases.
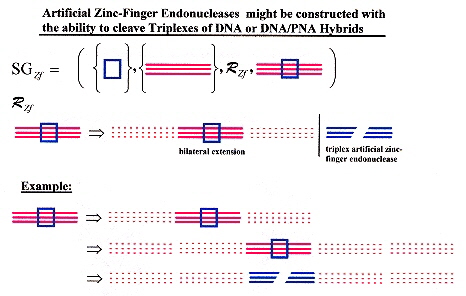
a "Restriction Endonucleases", Edited by A. Pingoud, Springer, New York, 2004, p. 13
"Zinc Finger Domains: From Predictions to Design", by J. M. Berg, Accounts of Chemical Research, 1995, 28, 14 - 19
Zinc finger proteins found in TFIIIA-5S RNA from Xenopus are of the form (Tyr, Phe)-X-Cys-X2-4-Cys-X3-Phe-X5-Leu-X2-His- X3,4-His-X2-6.

"Site-specific cleavage of DNA-RNA hybrids by zinc finger/FokI cleavage domain fusions", by Y-G. Kim, Y. Shi, J. M. Berg, S. Chandrasegaran, Gene, 1997, 203, 43 - 49
In zinc finger protein-DNA complexes, one strand of DNA is bound much more than the other strand. However, chimeric endonucleases can be extended to DNA-RNA hybrids. The modular structure of FokI endonucleases suggested that it might be feasible to construct chimeric restriction enzymes with novel sequence specificities by linking other DNA-binding proteins to the cleavage domain of FokI. To be explicit, zinc finger protein binding can be linked to the cleavage domain of FokI to create 'artificial' nucleases with designed cleavage sites.
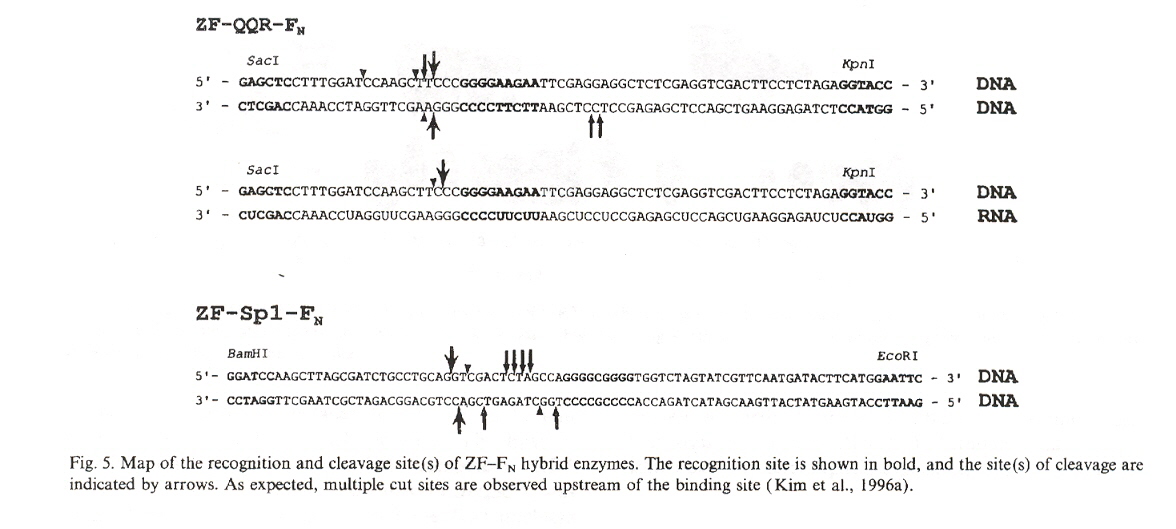
"Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain", by Y-G. Kim, J. Cha, S. Chandrasegaran, Proceedings of the National Academy of Sciences U.S.A., Feb. 1996, 93, 1156 - 1160
It is pointed out in this paper that the chimeric endonucleases created cleave double-stranded DNA at many cleavage sites, and that each one of these sites in a map vary by observed percentages.
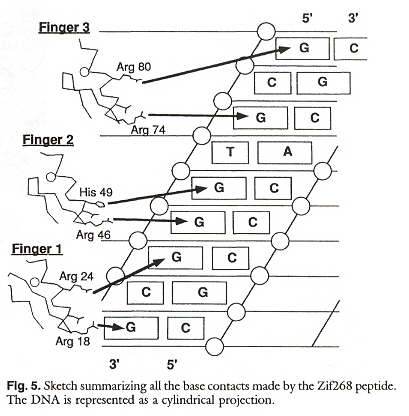
"Zinc Finger-DNA Recognition: Crystal Structure of a Zif268-DNA Complex at 2.1 Å", by N. P. Pavletich, C. O. Pabo, Science, 1991, 252, 809 - 817
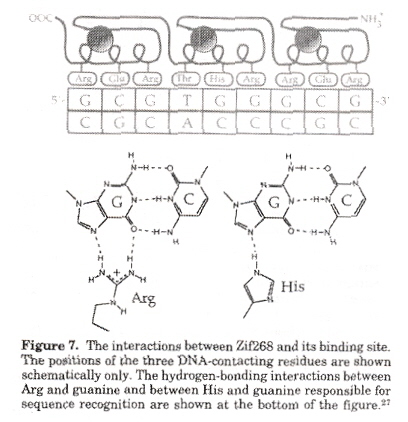
In this paper, it is pointed out that most of the contacts at the nucleotide binding sites are with guanine.
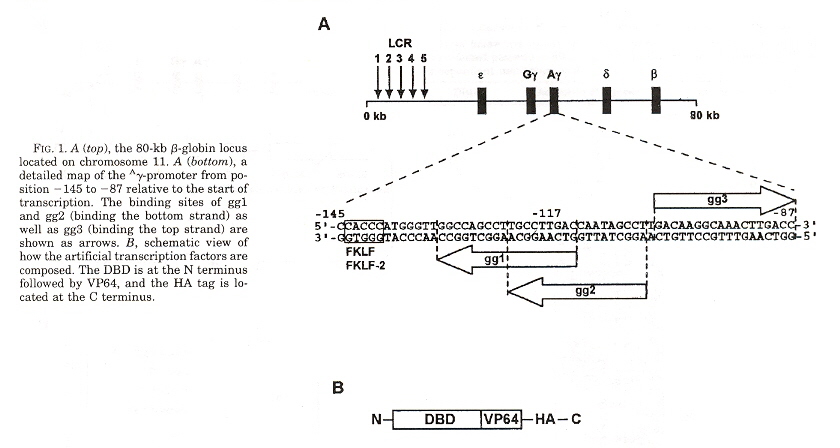
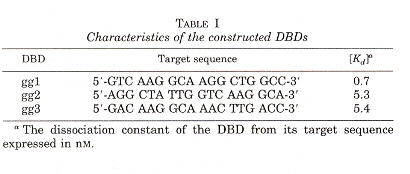
"Exploring Strategies for the Design of Artificial Transcription Factors: Targetting Sites Proximal to Known Regulatory Regions for the Induction of γ-Globin Expression and the Treatment of Sickle Cell Disease", by T. Gräslund, X. Li, L. Magnenat, M. Popkov, C. F. Barbas III, Journal of Biological Chemistry, Feb. 4 2005, 280, 5, 3707 - 3714
Zinc finger-based transcriptional activators designed to target -117-position of the λ-globin promotor. Three DNA binding domain λ-globin promotors were designed, called gg1, gg2, and gg3. The λ-globin content of cells expressing gg1-vp64-HA showed up to 16-fold higher levels of fetal hemoglobin.
"Zinc finger proteins: getting a grip on RNA", by R. S. Brown, Current Opinion in Structural Biology, Feb. 2005, 15, 1, 94 - 98
Zinc fingers not only bind to DNA, but also bind to ssRNA and dsRNA! Examples include zinc finger binding to viral proteins, plant proteins, and parasites, including mitochondrial RET1 uridylyl transferase, etc.
"Artificial Zinc Finger Peptides: A Promising Tool in Biotechnology and Medicine", by N. Corbi, V. Libri, C. Passananti, Handbook of Experimental Pharmacology (HEP), 2004, 166, 491 - 507
A recognition code for zinc finger Zif268 is in the figure below. Experimental methods are based upon parallel selection or sequential selection. In parallel selection, the first and third zinc fingers are fixed (anchor fingers) for correct positioning on the target site, while the second zinc finger is randomized. For sequential selection, zinc fingers are serially selected adding one finger at a time, allowing context-dependent interactions between zinc fingers and subsites.
Polydactyl zinc fingers with more than three fingers are also investigated, including as many as six fingers, targeting 18 base pairs.
Zif268 Recognition Code
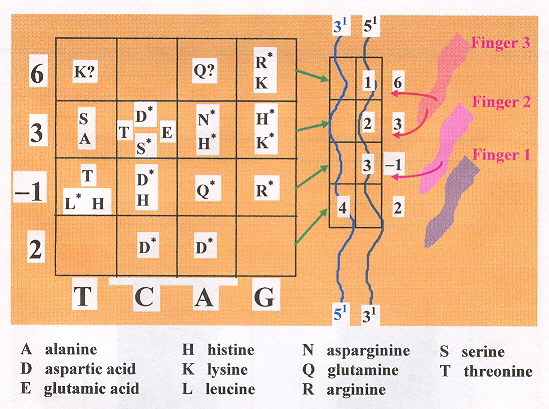
Amino acid residues that arise recurrently from phage display selections are in bold, asterisks indicate interactions observed in structural studies. Uncertain correspondences are indicated by a question mark. Poorly defined correspondences are left blank.
"Effects of different zinc finger transcription factors on genomic targets", by L. W. Neuteboom, B. I. Lindhout, I. L. Saman, P. J. J. Hooykaas, B. J. van der Zaal, Biochemical and Biophysical Research Communications, Jan. 6 2006, 339, 1, 263 - 270
Polydactyl Zinc Fingers (PZF) such as two three-finger units (2 × 3F), or three two-finger units (3 × 2F), both of which are 6F, but 4F, 5F, etc. are also discussed. Each zinc finger is joined by a linker (a short sequence of polypeptides used to control the distance between each finger). Various researchers have used different linkers. Linkers can be constructed by using the appropriate codon for each amino acid, the base sequences inserted into a plasmid. Linkers used include the following.
"Specificity Changes in the Evolution of Type II Restriction Endonucleases", by V. Pingoud, A. Sudina, H. Geyer, J. M. Bujnicki, R. Lurz, G. Läder, R. Morgan, E. Kubareva, A. Pingoud, Journal of Biological Chemistry, Feb. 11 2005, 280, 6, 4289 - 4298
There are a number of Type II restriction endonucleases that might appear to be entirely distinct but upon examination are quite similar. The Type II restriction endonuclease MboI with ↓GATC is shown to most likely have a common evolutionary origin as the following Type II restriction endonucleases (note that these restriction endonucleases have different "specificities" or types of cleavage).
| SsoII | ↓CCNGG | N = A, C, G, T |
| PspGI | ↓CCWGG | W = A, T |
| EcoRII | ↓CCWGG | W = A, T |
| NgoMIV | G↓CCGGC | |
| Cfr10I | R↓CCGGY | R = Pu (purine), Y = Py (pyrimidine) |
There are a very few Type II restriction endonucleases that cleave the same sequence at the same
position (see below). Neoschizomers recognize the same sequence of bases, but cleave at
different positions. Thus AatII recognizes and cleaves at GACGT↓C, while ZraI recognizes
GAC↓GTC. MTases add methyl groups to bases. While restriction endonucleases cleave
both strands of dsDNA, nicking enzymes cleave only one strand (either top or bottom),
Thus:
Nt.Bpu10I 51—CC↓TNAGC
31—GGANTCG
Nb.Bpu10I 51—CCTNAGC
31—GGANT↑CG
| EcoRI | G↓AATTC | |
| RsrI | G↓AATTC | |
| MthTI | GG↓CC | |
| NgoPII | GG↓CC | |
| XmaI | C↓CCGGG | |
| CfrI | C↓CCGGG | |
| Cfr10I | R↓CCGGY | R = Pu (purine), Y = Py (pyrimidine) |
| Bse634I | R↓CCGGY | R = Pu (purine), Y = Py (pyrimidine) |
However, there are restriction endonucleases that cleave in partially related sequences:
| EcoRI | G↓AATTC | |
| MunI | C↓AATTG | |
| SsoII | ↓CCWGG | W = A, T |
| PspGI | ↓CCNGG | N = A, C, G, T |
As a consequence, the prevailing view (until 1995) was that restriction enzymes are not related evolutionarily. The view changed as it became clear that restriction enzymes have a very similar structure: four-stranded β-sheet flanked by α-helices and the characteristic PD...(D/E)XK motif active site. The current view is that this family of restriction enzymes evolved by divergent evolutiona.
a "Restriction Endonucleases", Springer-Verlag, 2004, A. Pingoud (ed), 63 - 93 (J. M. Bujnicki).

"Review; Type II restriction endonucleases: structure and mechanism", by A. Pingoud, M. Fuxreiter, V. Pingoud, W. Wende, Cellular and Molecular Life Sciences, March 2005, 62, 6, 685 - 707
This paper is an excellent review of restriction endonucleases (although there is another very good referencea). The various restriction endonucleases include the following Types.
To get an idea of the distribution of the more than 3707 types of restriction endonucleases:
Type II restriction endonucleases recognize palindromic base pair sequences 4 to 8 in length (hence Type IIP, "P" for palindromic), and cleave within the sequence in both strands. "Sticky end" overhangs or "blunt" ends result after cleavage.
Binding to a binding site means opening up the DNA. Binding can take place in two ways:
a "Restriction Endonucleases", Edited by A. Pingoud, Springer, New York, 2004.
"Engineered Zinc Finger Proteins that Respond to DNA Modification by HaeIII and HhaI Methyltransferase Enzymes", by M. Isalan, Y. Choo, Journal of Molecular Biology, Jan. 21 2000, 295, 3, 471 - 477
Cytosine modified by a zinc finger into 5-methylcytosine (5-mC) is very common. One example is that an infecting virus may not have a methylated cytosine, and its DNA gets cleaved by a restriction endonuclease. This paper reports a zinc finger engineered to discriminate and bind only when the cytosine is methylated by a methyltransferase. Methyltransferases M.HaeIII and M.Hha I will methylate the cytosines in bold in: GGCCCGGCG and GCGCCGGCG.
Zinc Fingers used with a Methytransferase
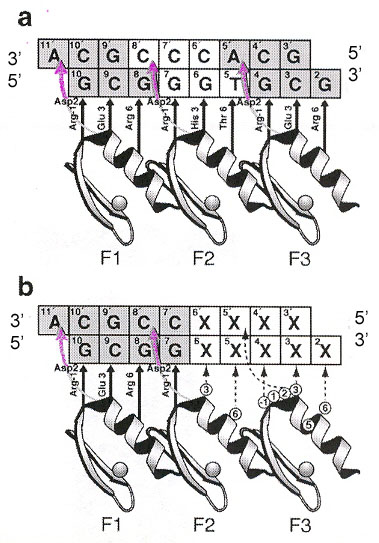
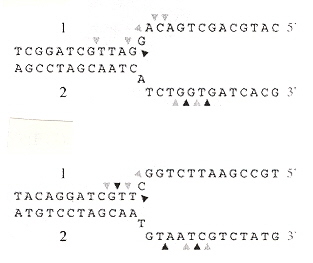
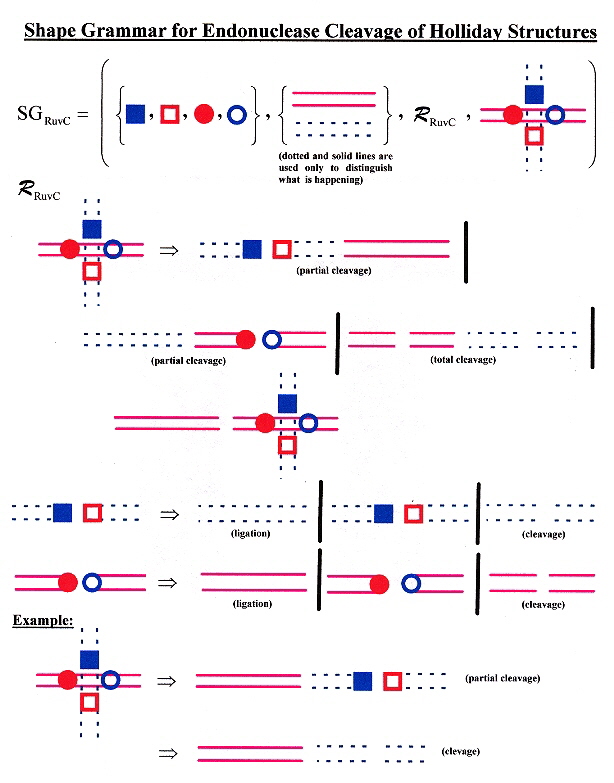
"Evolution of a phage RuvC endonuclease for resolution of both Holliday and branched DNA junctions", by F. A. Curtis, P. Reed, G. J. Sharples, Molecular Microbiology, March 2005, 55, 5, 1332 - 1345
Enzyme 67RuvC cleaves both Holliday structures as well as branched (fork) junctions.
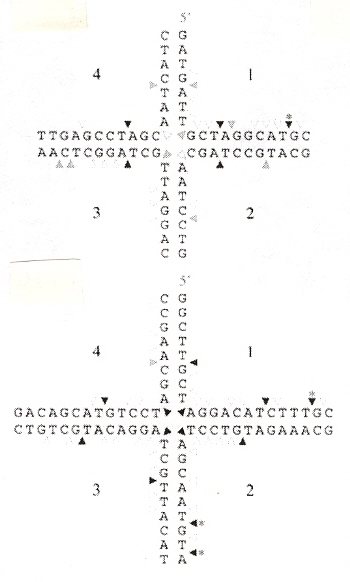
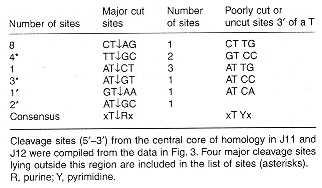
Shape Grammar for RuvC Holliday Structure Cleavage
"The Artemis: DNA-PKcs endonuclease cleaves DNA loops, flaps and gaps", by Y. Ma, K. Schwarz, M. R. Lieber, DNA Repair, July 12, 2005, 4, 7, 845 - 851
Interesting mechanisms of DNA-dependent protein kinase to correct DNA damage are proposed. Repair mechanisms include cleavage of overhangs, ligation and removal of flaps to get gaps followed by stem-loop formation, then cleavage of loops and flaps with ligations to get repaired double stranded DNA.
"A tRNATrp Intron Endonuclease from Halobacterium volcanii: UNIQUE SUBSTRATE RECOGNITION PROPERTIES", by L. D. Thompson, C. J. Daniels, The Journal of Biological Chemistry, Dec. 5 1988, 263, 34, 17951 - 17959
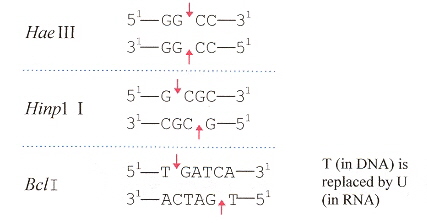
Different cleavage patterns from different restriction endonucleases are defined in the first figure. Part B of the second figure (see footnote "a") shows how an intron from DNA is excised from the corresponding DNA, to obtain the form of an RNA secondary conformation seen in Part A of this figure. The third figure takes a complex tRNATrp and a RNA restriction endonuclease that cleaves at two places just like Hinp1 I and deletes the RNA intron between both cleavage sites, then ligates to obtain the fourth figure. From the point of view of evolution, studying Desulfurococcus mobils b, it is possible that tRNA and rRNA introns in eukaryotes may have a common archaebacterial origin.
Restriction Endonucleases
Intron Excision
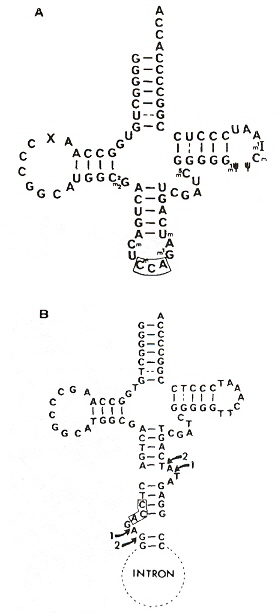
H. mediterranaei tRNATrp Intron Excision
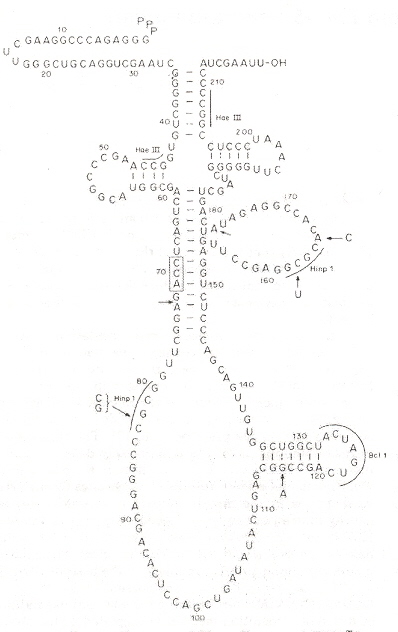
Intron Excised

a "Transcription and Excision of a large Intron in tRNATrp Gene of an Archaebacterium, Halobacterium volcanii", by C. J. Daniels, R. Gupta, W. F. Doolittle, The Journal of Biological Chemistry, Dec. 5 1988, 263, 34, 17951 - 17959
b "Novel Splicing Mechanism for Ribosomal RNA Intron in the Archaebacterium Desulfurococcus mobilis", by J. Kjems, R. A. Garrett, Cell, Aug. 26 1988, 54, 693 - 703
As the figure below shows, a primer with an OH functional group couples (Watson-Crick compatible), starting at the 3' end of a ssDNA strand, using dATP, dCTP, dGTP, or dTTP as required), and step-wise pairs agaist the ssDNA template.
Polymerase Mechanism

"Exploring the Recognition of Quadruplex DNA by an Engineered Cys2-His2 Zinc Finger Protein", by S. Ladame, J. A. Schouten, J. Roldan, J. E. Redman, S. Neidle, S. Balasubramanian, Biochemistry, 2006, 45, 1393 - 1399
Engineered zinc finger protein Gq1 binds with intramolecular G-quadruplex 5'–(GGTTAG)5–3'. Any one finger of Gq1 can be replaced by Zif268 without significant loss of quadruplex affinity or quadruplex discrimination.
"Eukaryotic topoisomerase II cleavage of parallel stranded DNA tetraplexes", by I. K. Chung, V. B. Mehta, J. R. Spitzner, M. T. Muller, Nucleic Acids Research, April 25 1992, 20, 8, 1973 - 1977
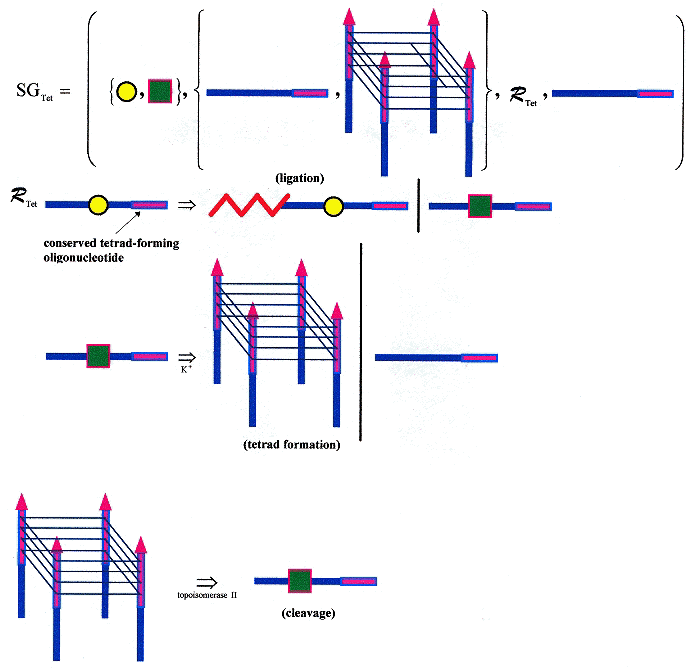
DNA cleavage by topoisomerase II is blocked by DNA triplex formation. ssDNA oligonucleotide "M" is not cleaved by topoisomerase II, but four copies of M, forming the oligonucleotide "M4" as a quadruplex is cleaved! "M" as dsDNA is not cleaved. Thus ssDNA can fold into a tetraplex, then be cleaved, and using single strand ligation, the process can be repeated. Obviously a language can be created for this process. While the cleavage by topoisomerase II takes place in vitro, such a language may not take place in vivo, but could nevertheless be useful to chemists workin in nanotechnology (for example). Clearly, different ssDNS with the same (conserved) tetrad forming terminals could be used to construct automata, as well. This is just one more example that goes well beyond splicing systems and dominos.
Below is a shape grammar that supports ligation with a conserved ssDNA tetrad-forming terminal. Ligation allows an infinite number of ssDNA to be generated, such that each strand can participate in tetrad formation. Using K+, a tetrad can form from ssDNA; using topoisomerase II, tetrads can be cleaved. Various kinds of tetrads can form, with different configurations.
Tetrad Forming/Cleaving/Ligating Shape Grammar
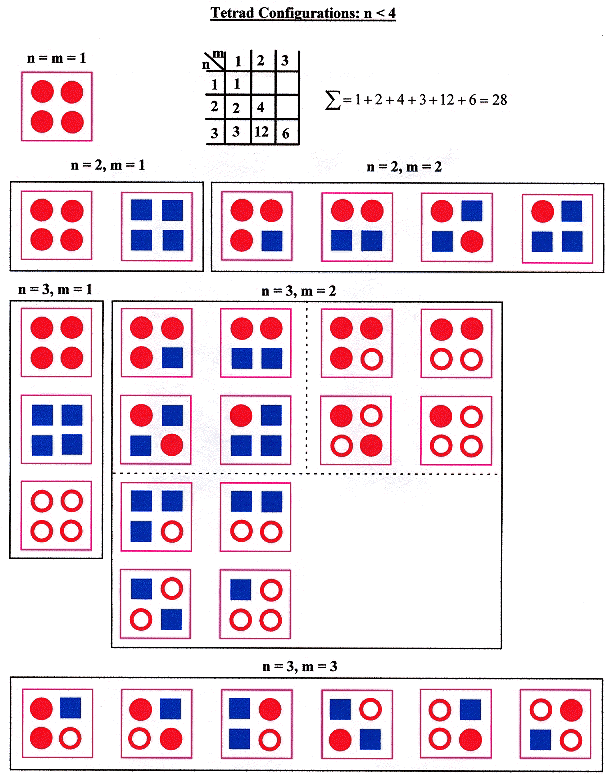
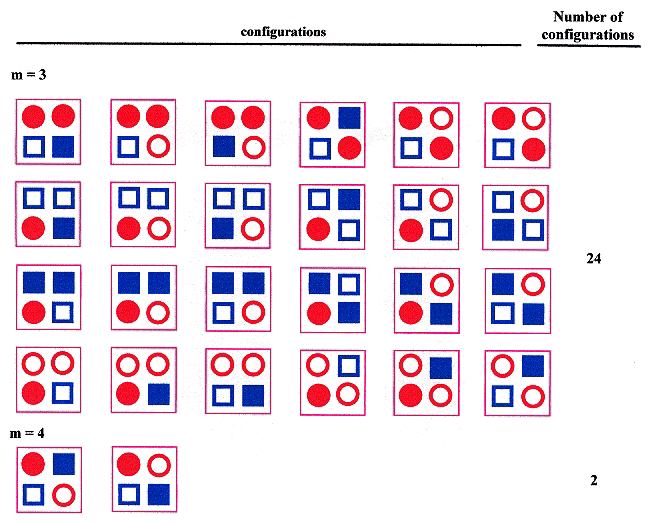
Given that n is the number of types of ssDNA, and m is the number of types of sDNA in a tetrad, then 1 ≤ m ≤ n, and one should obtain the following result.
Tetrad types and Configurations, n < 4
Tetrad types and Configurations, n = 4
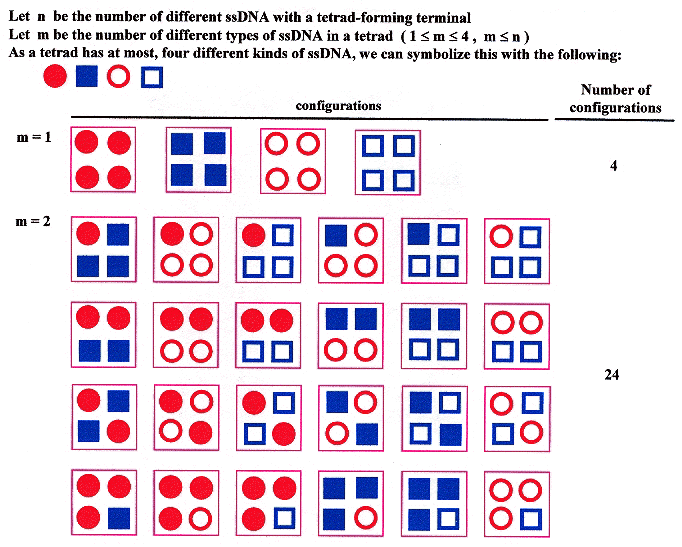
Tetrad types and Configurations, n = 4 (continued)
© Matthew Simon, 2005 - 2017