DNA Nano-Self-Assembly
"Design and Characterization of Programmable DNA Nanotubes", by P. W. K. Rothemund, A. Ekani-Nkodo, N. Papadakis, A. Kumar, D. K. Fygenson, E. Winfree, Journal of the American Chemical Society, 2004, 126, 163444 - 16352
Tiles (segments of double-helix DNA with sticky-ends) may be "programmed" by selecting different sticky-ends. It was observed that the "sheets" of (2-dimensional) DNA could fold over into nanotubes. It is interesting that a shape grammar can be used in this work.
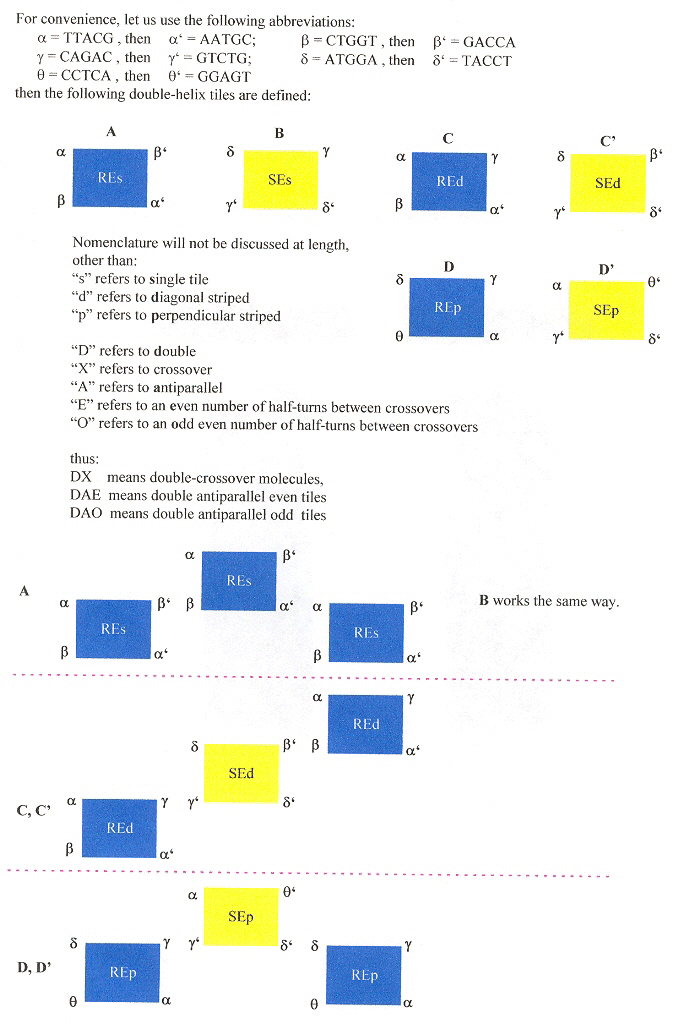
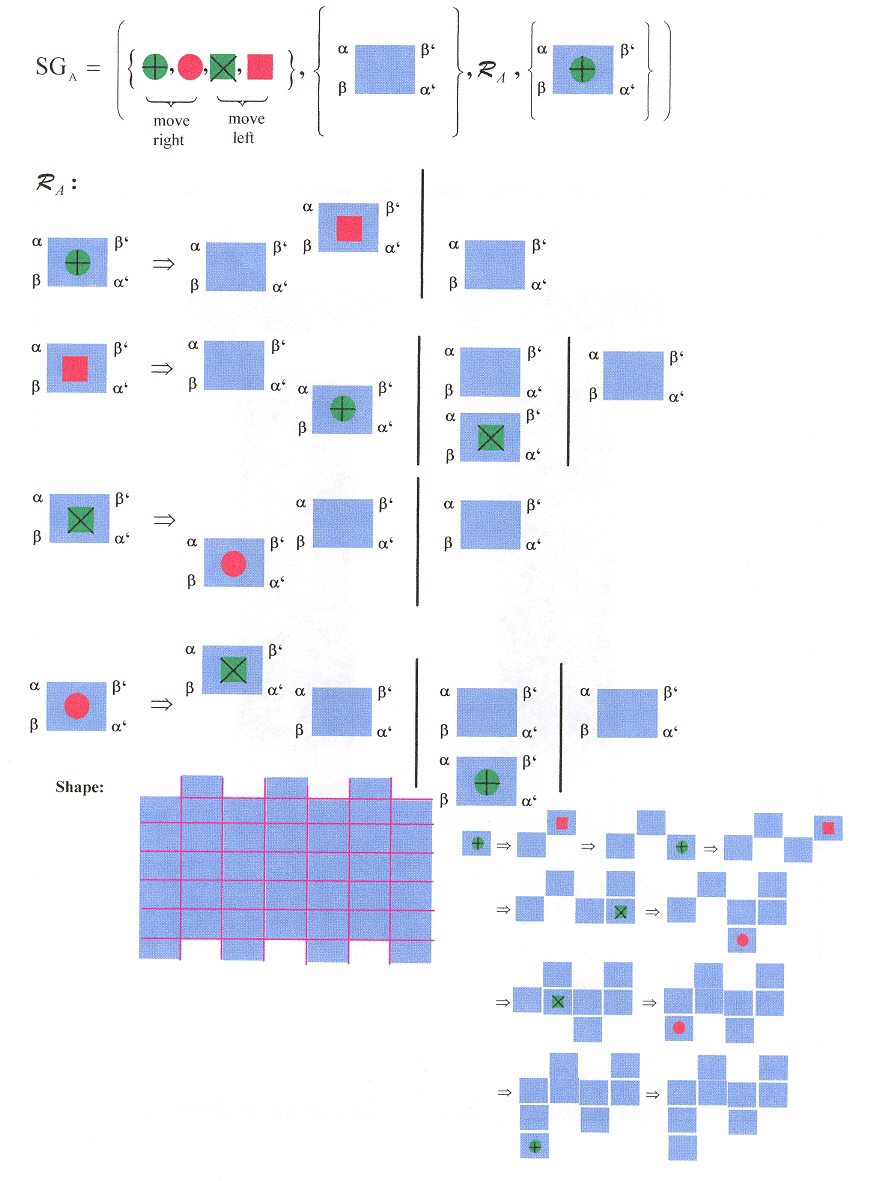
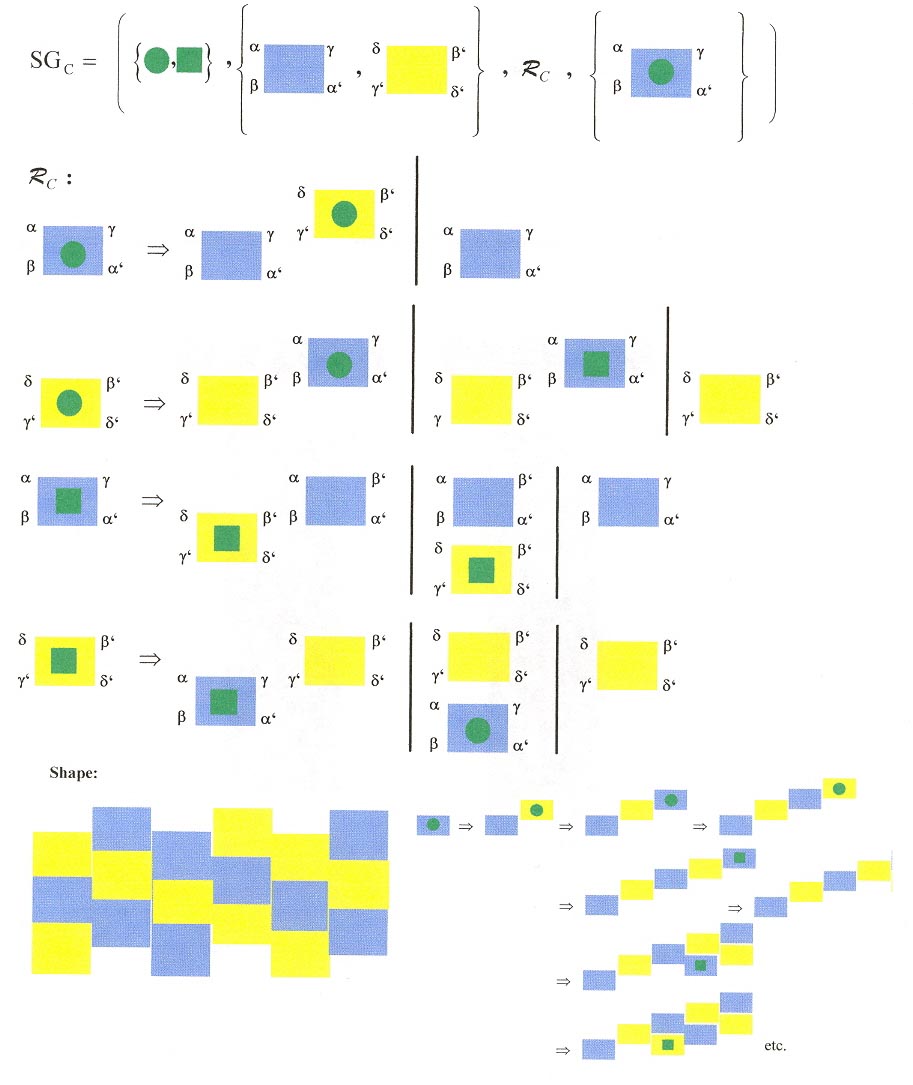
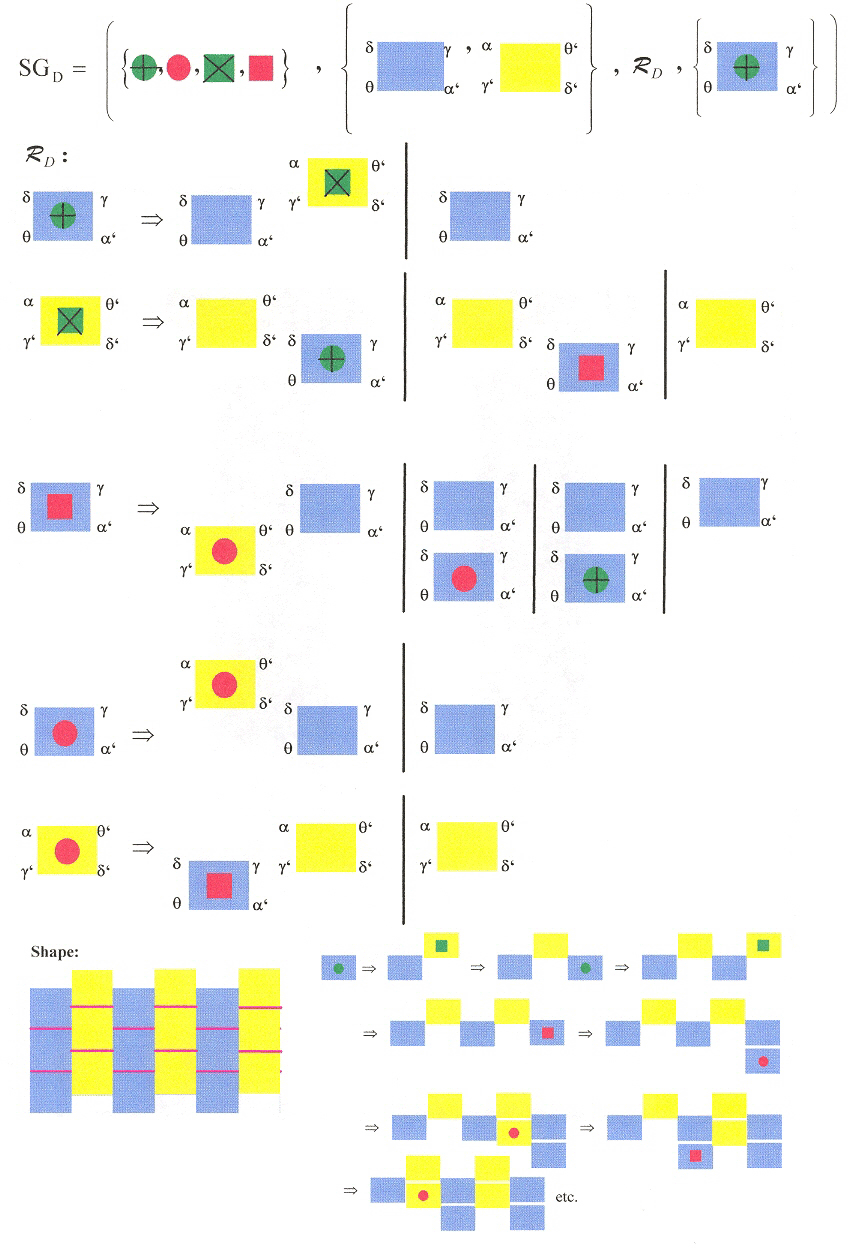
Three shape grammars as examples:
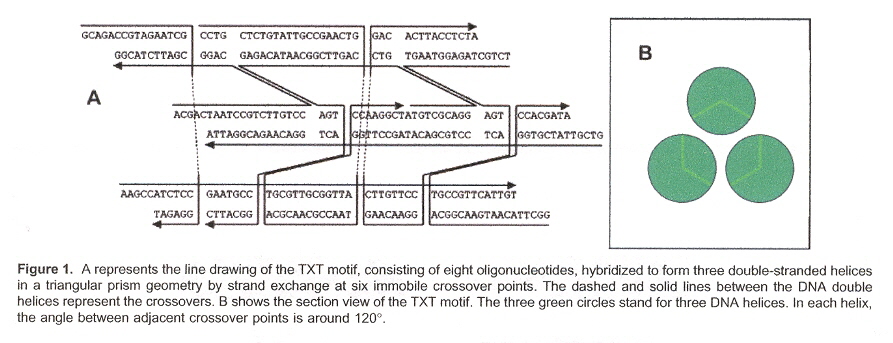
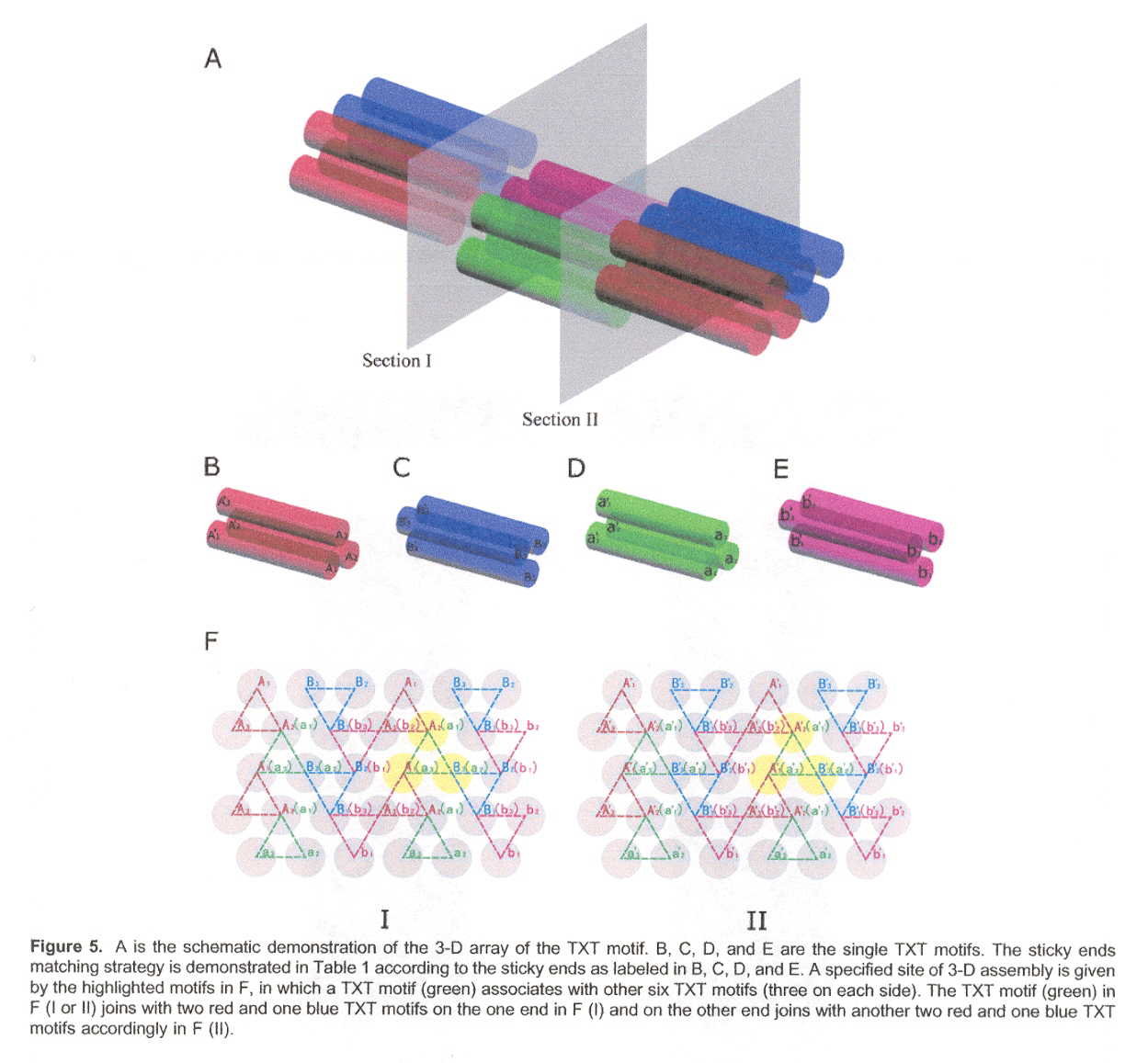
"A New Triple Crossover Triangle (TXT) Motif for DNA Self-Assembly", by B. Wei, Y. Mi, Biomacromolecules, Sept. 2005, 6, 5, 2528 - 2532
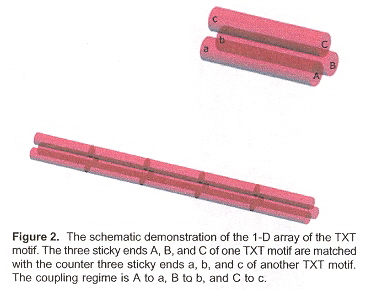
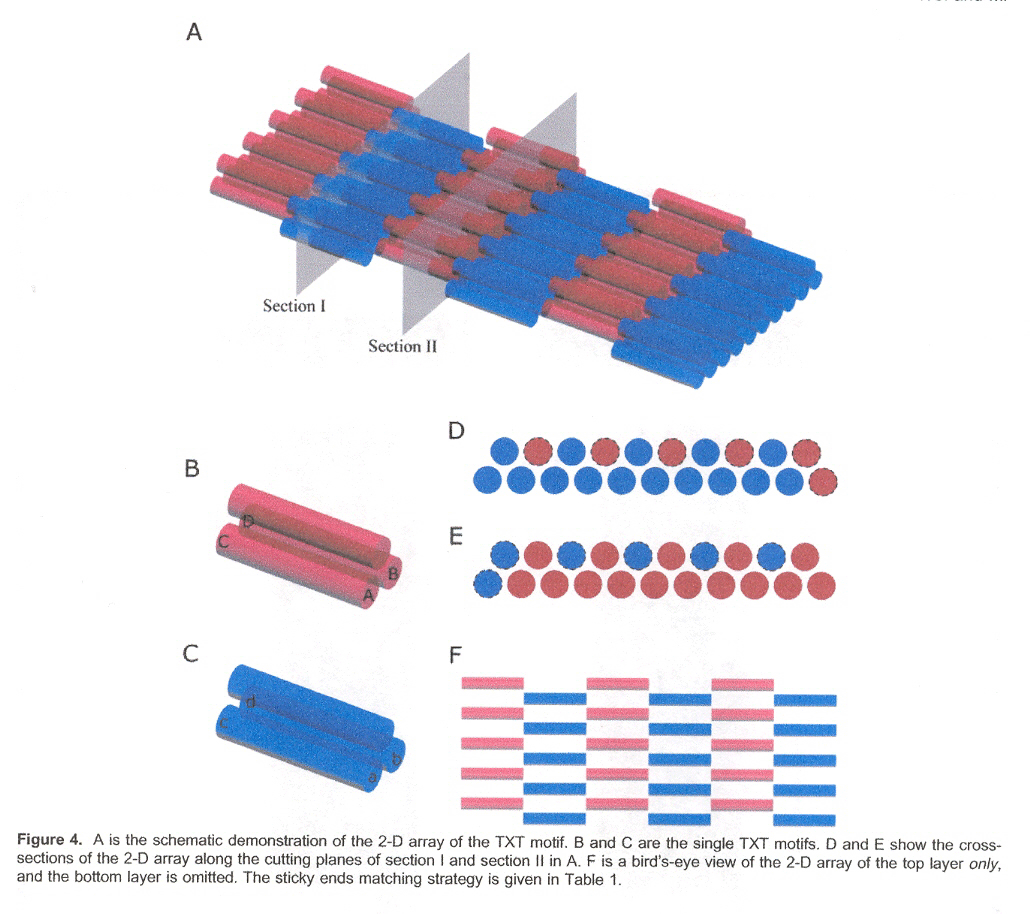
This paper discusses how 1-, 2- and (especially) 3-dimensional self-assembling structures can be designed (using sticky end selection in different associations)a. The desired associations are seen in Table 1, and for 3-dimensional structures, the associations are given in Figures B through F.
aNote: See "Construction, Analysis, Ligation, and Self-Assembly of DNA Triple Crossover Compexes", by T. H. LaBean, H. Yan, J. Kopatsch, F. Liu, E. Winfree, J. H. Reif, N. C. Seeman, Journal of the American Chemical Society, 2000, 122, 1848 - 1860
Shape grammars could be developed for all three dimensions, but they will not be done here.
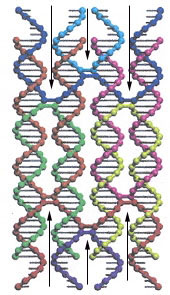
"Self-Assembly of DNA Double-Double Crossover Complexes into High-Density, Doubly Connected, Planar Structures", by D. Reishus, B. Shaw, Y. Brun, N. Chelyapov, L. Adleman, Journal of the American Chemical Society, Dec. 21 2005, 127, 50, 17590 - 17591
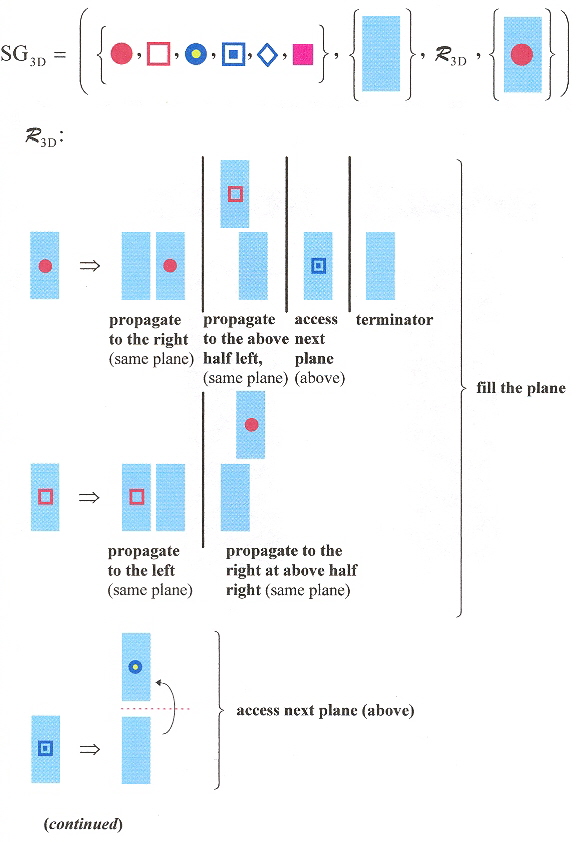
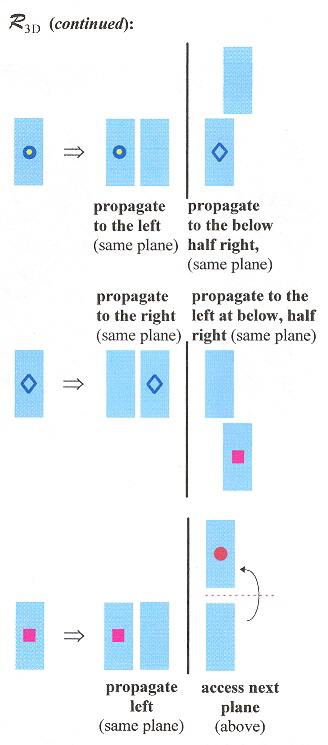
Double-double crossover (DDX) composed of four dsDNA helices, each adjacent dsDNA strand joined by a pair of sticky-ends (for a total of 6 sticky-end connections - see the "arrows" in the first figure below) is described in this paper. Planar structures are observed experimentally, but self-assembly into three-dimensional structures is also possible. A shape grammar for 3-D structures is provided. This grammar assembles DDX structures in "on-end" (vertical) rod-like form, but can also be viewd as planar sheets in 3D form.
DNA Double-Double Crossover (DDX)
Four adjacent (vertcical) dsDNA are doubly-linked with sticky-ends
|
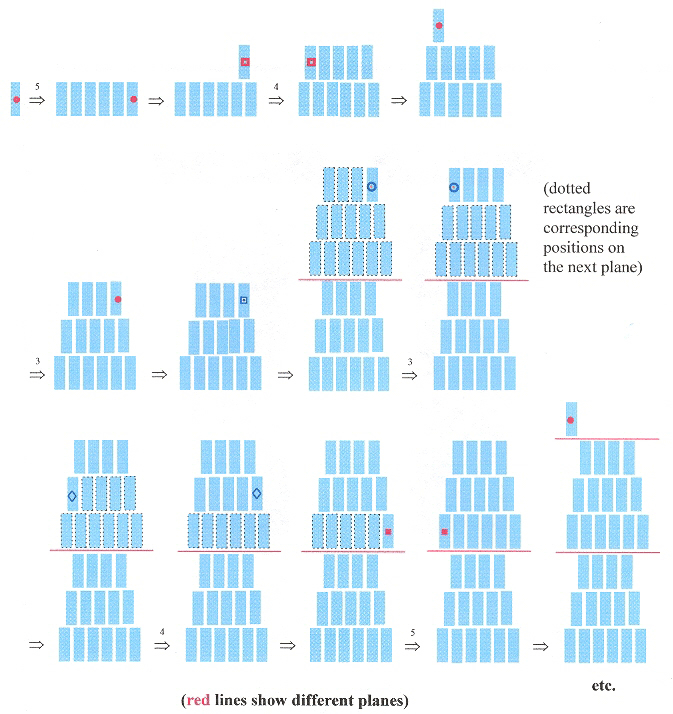
3–D Shape Grammar
|
3–D Shape Grammar (continued )
|
Sample generation of SG3D
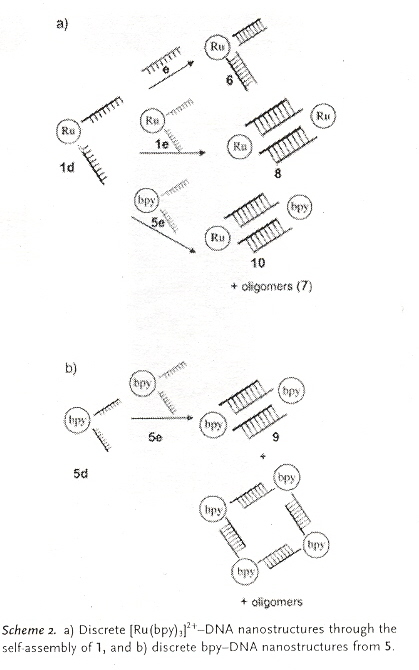
"Self-Assembly of Cyclic Metal–DNA Nanostructures using Ruthenium Tris(bipyridine)-Branched Oligonucleotides", by D. Mitra, N. Di Cesare, H. F. Sleiman, Angewandte Chemie International Edition, Nov. 2 2004, 43, 43, 5804 - 5808
Nanostructures containing short DNA duplexes as arms with transition metal vertexes as centers are formed. Specifically-addressable transition metals support properties such as luminescence and redox (oxidation and reduction).
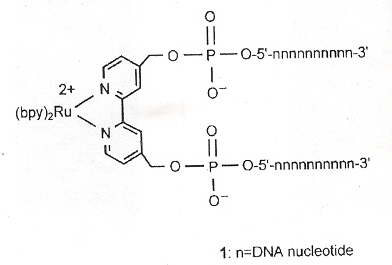
Specifically, ruthenium (II)–DNA forms a rigid complex with two DNA strands as seen below. These complexes self-assemble into cyclic bipyridine/double-stranded DNA structure.
"Nucleic Acid Nanotechnology", H. Yan, Science, December 17, 2004, 306, 5704, 2048 - 2049
Applications of DNA and RNA to nanotechnology similar to those discussed in the Appendix (Shape Grammars) in "Emergent Computation: Emphasizing Bioinformatics".
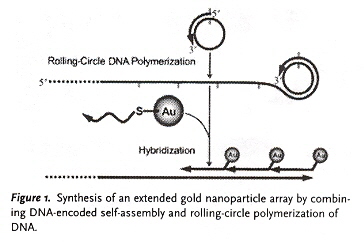
"DNA-Encoded Self-Assembly of Gold Nanoparticles into One-Dimensional Arrays", by Z. Demg, Y. Tian, S-H. Lee, A. E. Ribbe, C. Mao, Angewandte Chemie International Edition, June 6 2005, 44, 23, 3582 - 3585
The authors construct a combined mixture of Gold nanoparticles (AuNPs) with DNA 1 (AuDNA1). These mixes of AuNPS/AuDNA1 are then polymerized using rolling-circle polymerization to form large one-dimensional arrays of AuNP.
-
(review rolling-circle replication in mtDNA.html#ROLCR)
- Rolling-Circles and Codons
- Rolling-Circles and Plasmids
Rolling-Circle DNA Self-Assembly of AuNP
DNA Nano-Quadruplex-Self-Assembly
"Guanine Quartet Networks Stabilized by Cooperative Hydrogen Bonds", by R. Otero, M. Schöck, L. M. Molina, E. Lægsgaard, I. Stensgaard, B. Hammer, F. Besenbacher, Angewandte Chemie International Edition, 2005, 44, 15, 2270 - 2275
Guainine deposited under ultraclean conditions on inert Au(111) substrate will self-assemble (with hydrogen bonding) into G-quartets (even in the absence of the DNA backbone as well as the absence of metal ions in solution). Using Scanning Tunnelling Microscopy (STM), each unit cell is composed of four G islands. This phenomenon is favored by kinetics rather than by thermodynamics. Quantum Mechanical (QM) methods such as Hartree-Fock and Discrete Fourier Transforms (DFT) confirm a quadruplex structure. In particular, the quartet structure is about twice as strong as they are with the dimer, this extra bonding force being referred to as Resonance-Assisted Hydrogen Bonding or (RAHB). The intermolecular hydrogen-bonding with RAHB stabilization supports the observed blocks of two-dimensional self-assembled structures. A two-dimensional shape grammar is thus possible.
G-Island Quadruplex Shape Grammar
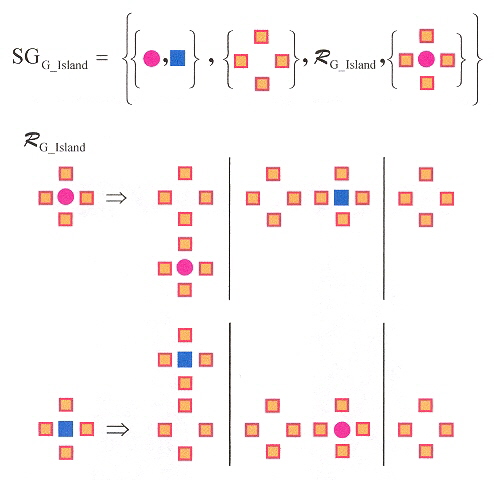
Generation of G-Island Quadruplex Network
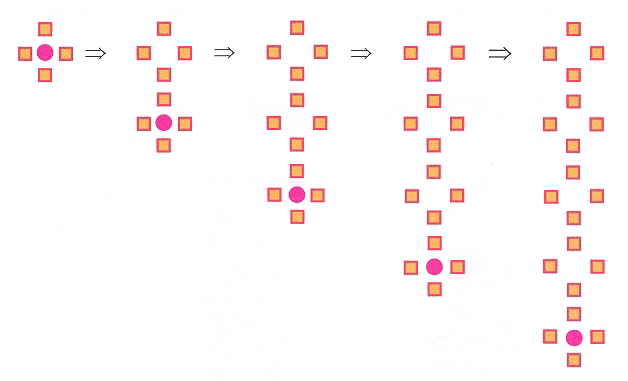
More G-Island Quadruplex Network Derivations
"DNA duplex–quadruplex exchange as a basis for a nanomolecular nachine", by P. Alberti, J.-L. Mergeny, Proceedings of the National Academy of Sciences USA, Feb. 18 2003, 100, 4, 1569 - 1573
Controlled transitions between a G-quadruplex structure folded and (compact) and its linear dsDNA form (as
proven by FRET UV emissions) supports the construction of a nanomachine. Transition from the linear dsDNA to
the quadruplex form in the presence of either Na+ or K+ is controlled by using "C-fuel"
or "G-fuel" (producing dsDNA GC duplex waste products). A 21-base d(GGGTTA)3 oligonucleotide is
used with fluorescence ("F") at the 5' end, and tetramethylrhodamine ("T") at the 3' end forming F21T. For
example, 3'–d(CCCAAT)3–5' is 21C "fuel", similarly for "G-Fuel".
G-quadruplex + C-fuel → dsDNA (linear)
dsDNA (linear) + G-fuel → G-quadruplex + GC-waste (thermodynamically more stable)
The distances between the ends of the two forms: linear dsDNA and the G-quadruplex cause a difference in UV emission. Cycling efficiency decreases as the waste product concentration increases. Eleven successive cycles have been achieved. The oscillation distances between the closed G-quadruplex and the open conformations have been calculted, as well as the precise force generated.
Back to TopDNA Nano-Wires
"Quantum Interference Device Made by DNA Templating of Superconducting Nanowires", D. S. Hopkins, D. Pekker, P. M. Goldbart, A. Bezryadin, Science, June 17, 2005, 308, 1762 - 1765
Applications of DNA to nanotechnology.
Back to TopRNA Nano-Self-Assembly
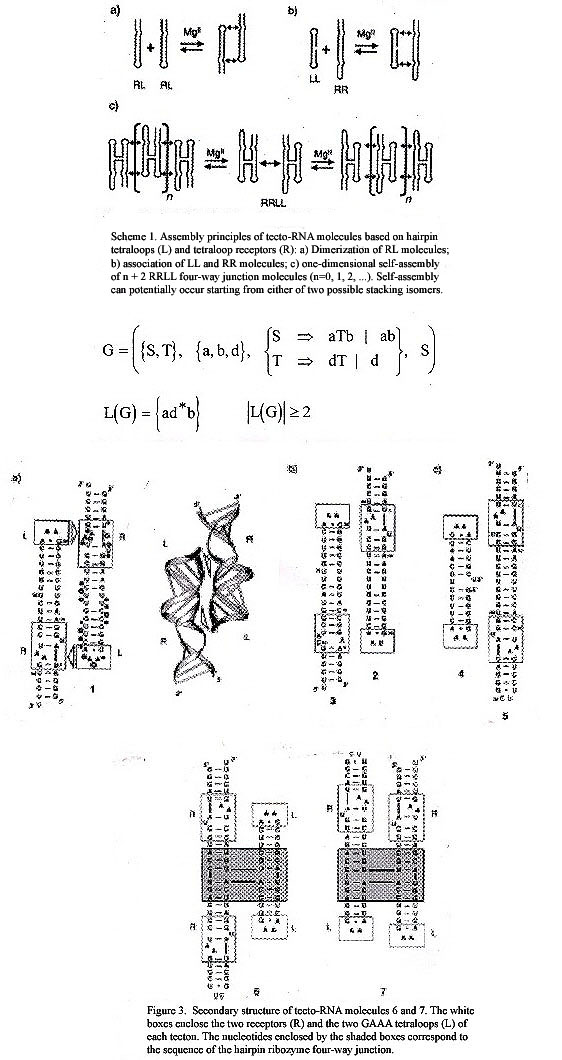
"Tecto-RNA: One-Dimensional Self-Assembly through Tertiary Interations", by L. Jaeger, N. B. Leontis, Angewandte Chemie International Edition, 2000, 39, 14, 2521 - 2524
Dimerization of RL RNA molecules where L refers to a tetraloop (of bases) and R refers to a tetraloop (of bases) receptor. Associations of LL with RR, then RRLL RNA linear self-assembling macromolecules. These are more than theoretical, as actual GAAA and GUAA tetraloop RNAs were found to be self-assembling.
"Building Programmable Jigsaw Puzzles with RNA", A. Chworos, I. Severcan, A. Y. Koyfman, P. Weinkam, E. Oroudjev, H. G. Hansma, L. Jaeger, Science, December 17, 2004, 306, 2068
Identical RNA subunits that the authors call "tectoRNA" can assemble into squares (se Fig. 1 A). With sticky ends, five types of tectosquares are possible (see Fig. 1 F), which can assemble into cis or trans configurations (see Fig. 1 E). Dimers, trimers, open tetramers, and closed tetramers can occur (see Fig. 2 A). As discussed in the Appendix to "Emergent Computation: Emphasizing Bioinformatics", shape grammars can be used to describe this process. Fig. 3 A, and Fig. 4 A show some experimental results found. Such materials can be useful in nanotechnology, and this is discussed in the following paper.
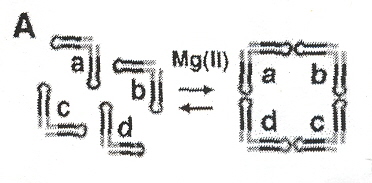
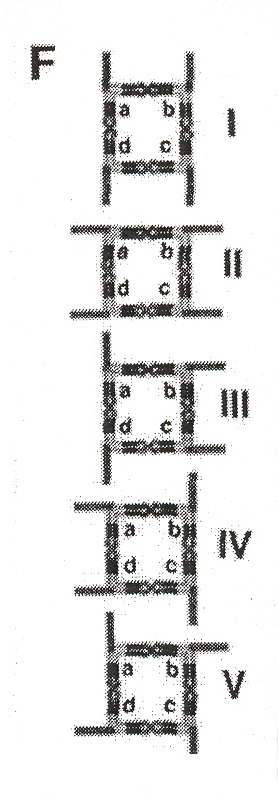
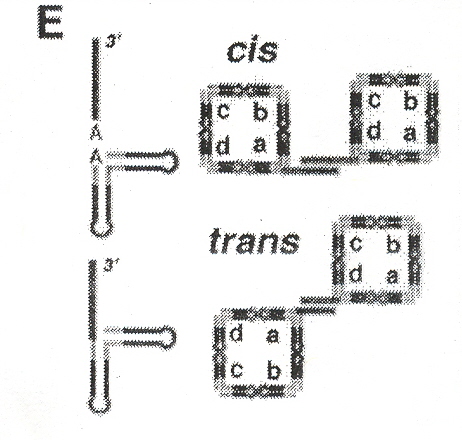
Figure 2A
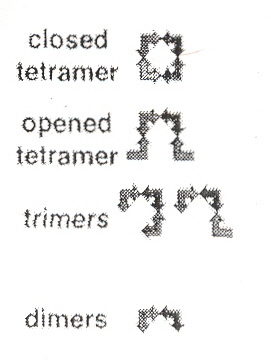
Figure 4A
Nano-Switches
"Individual Molecules of Dye-Labeled DNA Act as a Reversible Two-Color Switch upon Application of an Electric Field", S. S. White, L. Ying, S. Balasubramanian, D. Klenerman, Angewandte Chemie Int. Ed., Nov. 10 2004, 43, 44, 5926 - 5930
Using an electric field, rapid (less than 100 ms.), reversible DNA switching indicated by red and green fluorescence is described.
"DNA-Templated Three-Branched Nanostructures for Nanoelectronic Devices", by H. A. Becerril, R. M. Soltenberg, D. R. Wheeler, R. C. Davis, J. N. Harb, A. T. Woolley, Journal of the American Chemical Society, March 9 2005, 127, 9, 2828 - 2829
DNA templating to create discrete, three-branched metal nanostructures as precursors for three-terminal nanoelectronic devices. Three oligonucleotides that self-assemble using sticky-ends. The core is a ssDNA region hybridized with a biotinylated oligonucleotide that can be tagged with streptavidin. These were then metalized with copper as well as silver.
Assembly of a Labelled Three-Branched DNA Complex
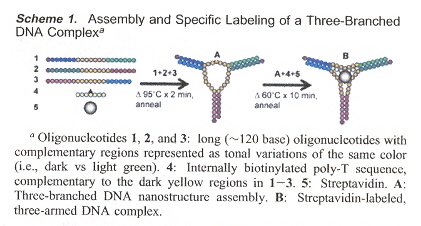
Nano State Machines
"Translation of DNA Signals into Polymer Assembly Instructions", by S. Liao, N. C. Seeman, Science, Dec. 17 2004, 306, 5704, 2072 - 2074
Two DNA molecules are viewed as devices "PX" and "JX2" with two states each. These devices are sequence dependent. The specific application is the positional control of polymer concatenation. Thus four ligated molecules are produced (below). This device could be used as a finite-state device.
- PX1 - PX2
- PX1 - JX22
- JX21 - PX2
- JX21 - JX22
"Using Gene Regulation to Program DNA-Based Molecular Devices", by W. U. Dittmer, S. Kempter, J. O. Rädler, F. C. Simmel, Small, 2005, 1, 7, 709 - 712
DNA tweezers are programmed. However, what is most interesting is how these tweezers are controlled. Genetic regulation of nano-tweezers is envisioned. The regulation protein LexA of the SOS regulon gene of E. coli, and the regulation protein LacI of the lac operon of E. coli are used to control the DNA tweezers in an artificial gene.
Back to TopNano Medical Applications
"Nanotechnology for the biologist", by S. E. McNeil, Journal of Leukocyte Biology, Sept. 2005, 78, 585 - 594
This very interesting overview of nanotechnology includes near-term applications including:
- Drug-delivery platforms
- Enhanced image-contrast agents (useful in MRI)
- Chip-based monitoring nanolabs
- Probes capable of tracking cells and molecules
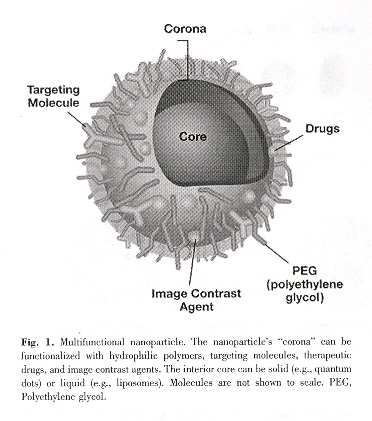
Combining multiple functions (above) into a single nanoscale entity, the opportunity to monitor and detect molecular and cellular changes due to diseases can also be reported. Such multifunctional devices are already in in vivo and in vitro testing. Other examples of nanotechnology:
- Dendrimers
- Liposomes
- Semiconducting quantum dots
Pharmaceuticals often fail to be of value due to problems with solubility. By chemically modifying the surfaces of nano-liposomes (core holds pharmaceuticals) with polyethylene glycol (PEG), called "pegylation", pharmaceuticals become encapsulated, effectively becoming soluble. Surface ligands convert these nanomaterials into biosensors, fluorescent tags, imaging agents, etc. (see Fig. 1). Due to their nanosizes, nanodevices can transit through blood vessels, cross the blood-brain barrier, stomach epithelium, avoid spleen filtration, avoid liver fenestrae and Kupffer sieve filtration. Charged particles can be located on dendrimers. As another example, silica-coated lipid micelles containing luteinizing hormone-release hormone (LH-RH) as a targetting agent can deliver ferric oxide quantum dots to LH-RH receptor positive cancer cells, and then after imaging, a rapidly oscillating magnetic field causes these dots to acts as projectiles and selectively kill only the cancer cells.
"Binding and Condensation of Plasmid DNA onto Functionalized Carbon Nanotubes: Toward the Construction of Nanotube-Based Gene Delivery Vectors", by R. Singh, D. Pantarotto, D. McCarthy, Oliver Chaloin, J. Hoebeke, C. D. Partidos, J-P. Briand, M. Prato, A. Bianco, K. Kostarelos, Journal of the American Chemical Society, March 30 2005, 127, 12, 4388 - 4396
This is a study concerned with the interaction of plasmid DNA with three kinds of functionalized Carbon nanotubes, the goal being to create a vector for plasmid DNA. Viral gene vectors can be immunogenic.
Single, Multi, and Single-Lysine walled nanotubes
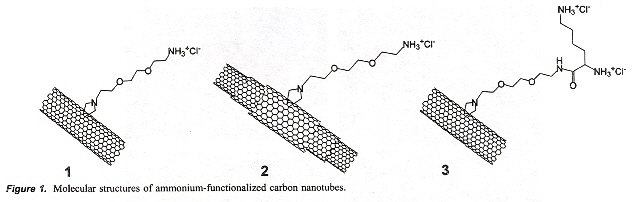
These functionalized carbon nanotubes are soluble, and plasmid pCMV-βgal were allowed to form complexes, tests were made at different concentrations of plasmid. Flourescent assays as well agarose gel electrophoresis show that functionalized carbon nanotubes can be used as plasmid DNA (especially, ssDNA) vectors.
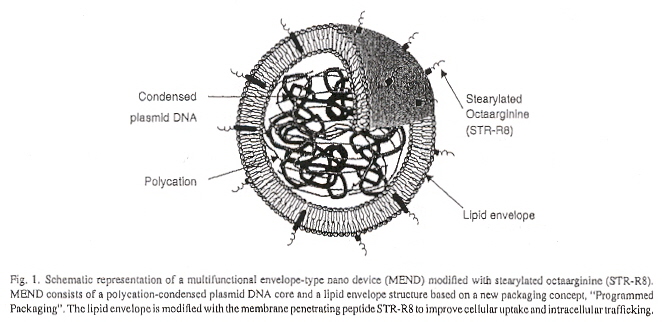
"A multifunctional envelope-type nano device for novel gene delivery of siRNA plasmids", by R. Moriguchi, K. Kogure, H. Akita, S. Futaki, M. Miyagishi, K. Taira, H. Harashima, International Journal of Pharmaceutics, Sept. 14 2005, 301, 1-2, 277 - 285
Gene delivery to the nucleus requires non-viral vectors to overcome
- Plasma membrane
- Endosomal Membrane
- Nuclear membrane
The multifunctional envelope-type nano device (MEND) succeeds as a non-viral vector. It is composed of an inner core of plasmid DNA with polycations (stabilizing cationic polymers, in this case, poly-L-lysine), a lipid envelope, with ocyaarginine (R8) membrane penetrating peptide. Cellular uptake of these nano-particles is by macropinocytosis (macrophages internalize the particles into macropinosomes). Lysosomal degradation is avoided and thus high transfection is reported.
MEND Nano-device Plasmid-DNA Vector
"PELGE Nanoparticles as New Carriers for the Delivery of Plasmid DNA", by X. Sun, Y-R. Duan, Q. He, J. Lu, Z-R. Zhang, Chemical Pharmacy Bulletin, 2005, 53, 6, 599 - 603
Nanoparticles can be an environmental hazard. This paper discusses biodegradable monomethoxy (poly-ethelene-glycol)-poly(lactide-co-glycolide)-monomethoxy(poly-ethelene-glycol) or PELGE plasmid DNA (pDNA) containing nanoparticles. The recent non-viral vector composed of nanoparticles using poly(DL-lactide-co-glycolide) or PLGA and polylacide PLA were of interest due to their biodegradability as well as the ability to protect DNA from degradation. PELGE was found to be effective.
"Interaction of fluorescent molecular rotors with blood plasma proteins", by W. J. Akers, J. M. Cupps, M. A. Haidekker, Biorheology, 2005, 42, 335 - 344
Many diseases have associated blood viscosity changes. Molecular rotors with flourescent markers can be used to measure blood viscosity. Such nanodevices rotate around a C=C double bond. As the viscosity increases, the rotation is inhibited favouring flouresence. The relationship between viscosity η and flourescent quantum yield Φ is lnΦ = x·lnη + C, where x is a dye dependent factor, and C is temperature dependent. This equation can be rearranged into η = (κ·I)ν where I replaces Φ and κ∝10–C and ν = x–1.
Molecular Rotor Molecules
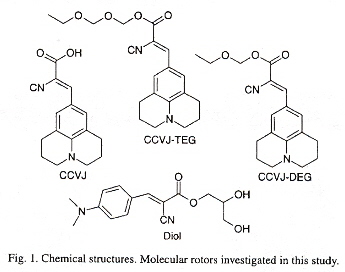
"A DNA-Based Machine That Can Cyclically Bind and Release Thrombin", by W. U. Dittmer, A. Reuter, F. C. Simmel, Angewandte Chemie International Edition, 2004, 43, 27, 3549 - 3553
The sequence 5'–GGTTGG TGT GGTTGGT–3' can fold into a two-plane G-quadruplex that binds
to α-thrombin protein. The basic function of this machine is as follows.
AP = Aptamer = 5'–TAAGTTCATCTC GGTTGG TGT GGTTGGT–3' (quadruplex)
AP-TB = AP bound to + α-thrombin
Q = 3'–GTTGCGGC ATTCAAGTAGAG CCAACC ACAC–5'
Thus AP-TB + Q → Q-AP-TB
Q-AP-TB → (linear dsDNA) Q-AP (G-quadruplex opens) + TB (α-thrombin is released)
Q-AP + R → (linear dsDNA) Q-R (waste) + AP (binds to α-thrombin to get AP-TB)
Nano Chemistry Applications
"The Chemistry of Organic Nanomaterials", by A. C. Grimsdale, K. Mällen, Angewandte Chemie International Edition, 2005, 44, 5592 - 5629
Defining "nano" to be in the 1 - 100nm range, the authors explicitly point out that the properties of nanoobjects often differ significantly from those of their corresponding bulk materials. The authors also point out that single molecules may be examined using:
The authors then discuss main classes of building units to construct nanomaterials:
- Phenylene-based conjugated polymers and oligomers (1-dimensional rigid-rods)
- Planar polycyclic aromatic hydrocarbons (2-dimensional PAHs)
- Rylene-based dyes
These materials can be thought of as subunits of graphene (a sheet of graphite). See Scheme 1.
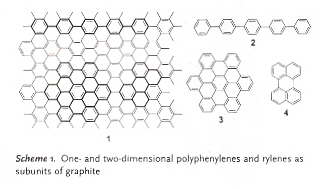
- Graphene
- Oligophenylene quinquephenyl
- Polyphenylene PAH
- Perylene
3-dimensional Polyphenylenes are called dendrimers, used as nanomaterial structural elements.
Rylene dyes include examples such as those in Scheme 10.
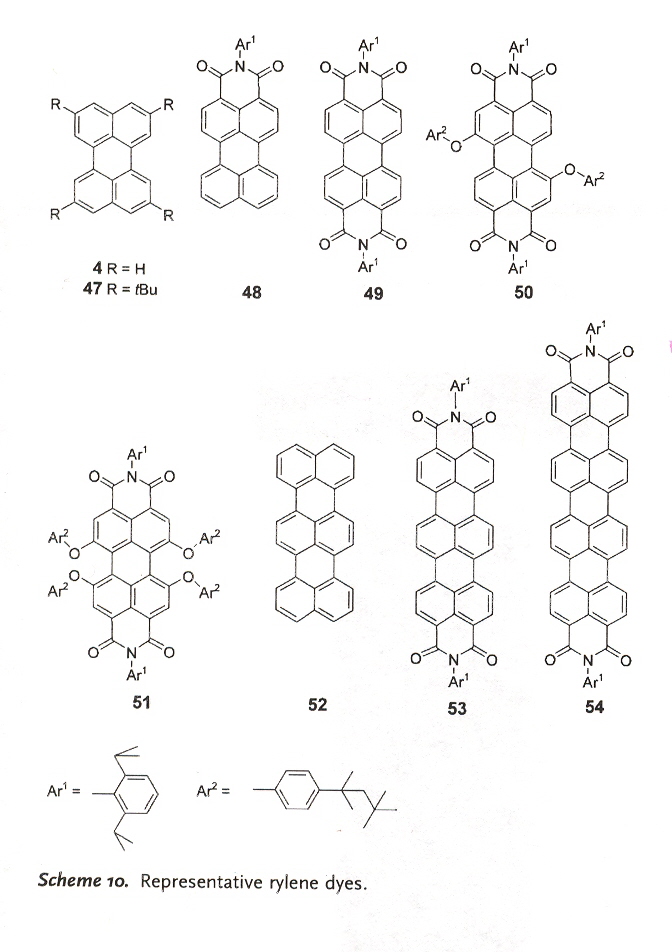
4 perylene blue-green 47 tetra-tert-butyl perylene blue 48 perylenemonoimide (PMI) yellow 49 perylenediimide (PDI) orange
50 diphenoxylated PDI orange 51 tetraphenoxylated PDI red 52 terrylene green 53 terrylenediimide (TDI) 650 nm 54 quaterrylene (QDI) 760 nm
Dendrimers are often used to deliver pharmaceuticals. Most dendrimers have flexible branches, easily deformed to bend back towards the center of the molecule. As a consequence, dendrimers are typically denser at the center as opposed to the periphery. Polyphenylene dendrimers are rigid, and they tend to be denser at the periphery as opposed to the center. Scheme 20 is an example of cis-trans isomerization that can be induced by light.
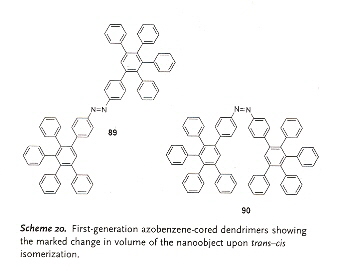
Rylene dyes may be attached to polyphenylene dendrimers in core positions, to result in emissive nanoparticles, see Scheme 27.
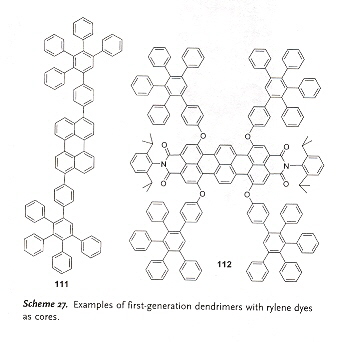
Alternatively, dendrimers with perylenemonoimide (PMI) chromophores (at the periphery) can be synthesized.
Using Shape Grammars, languages can easily be constructed to generate various dendrimers.
Back to TopDNA Nano-Motors
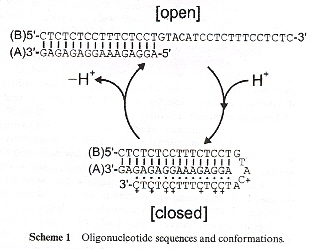
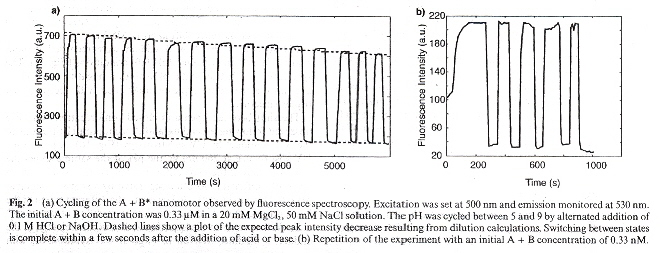
"The dynamic properties of an intramolecular transition from DNA duplex to cytosine-thymine motif triplex", by M. Brucale, G. Zuccheri, B. Samori, Organic and Biomolecular Chemistry, 2005, 3, 4, 575
By controlling the pH, an oligonucleotide sequence can change (in a short time) between a open DNA duplex or a closed DNA triplex. Thus this is a new kind of nanomotor. This was observed experimentally using fluorescent spectrophotometry.
"Molecular Motors: Synthetic DNA-Based Walkers Inspired by Kinesin", by T. R. Kelley, Angewandte Chemie International Edition, 2005, 44, 27, 4124 - 4127
Biological molecular motors such as flagella, muscles, cilia, etc. are expanded beyond rotary and linear motors to processive molecular motors which move across a surface. DNA based Kinesin, fueled by ATP, can "walk" along a microtubule, the direction being controlled. The "Shin-Pierce" walker can traverse a DNA track and return with full directional control. The "Tian & Mao" walker can traverse a circular track. The "Yan, Turberfield, Rief" walker also moves along a DNA track, but in a fashion similar to a "bucket brigade".
"A Free-Running DNA Motor Powered by a Nicking Enzyme", by J. Bath, S. J. Green, A. J. Turberfield, Angewandte Chemie International Edition, 2005, 44, 28, 4358 - 4361
A nano-technolgy application: a linear motor construced from DNA and a restriction endonuclease
(N.BbvCIB) moves a DNA cargo in discrete steps along a single stranded DNA track (composed of
"stators" Si). The cargo can bind (hybridize) to a stator. The restriction
endonuclease cleaves a single strand of DNA backbone (the track) thus introducing a nick.
The ability to create a nick is derived from a heterodimeric enzyme (BbvCI). The cargo is a
single nucleotide (which, once bound to a stator, the N.BbvCIB can then cut the stator, thereupon
the stator and cargo disassociate). The cargo cannot bind to stator S
dc1(t)/dt = – k1c1(t)
dc2(t)/dt = k1c1(t) – k2c2(t)
dc3(t)/dt = k2c2(t)
where ki is the rate of stepping along track stator Si to Si+1 and ci(t) is the fraction of cargo occupied at stator Si at time "t".
"Chiral Nanotechnology", by J. Zhang, M. T. Albelda, Y. Liu, J. W. Canary, Chirality, August 2005, 17, 404 - 420
Chiral nanotechnology includes nanotechnology in which chirality plays a useful role. 'Nano-chiral technology' is distinguished from 'chiral nanotechnology'. Nanoscale approaches to chiral technology include asymmetric synthesis and catalysis, enantioseparation, analytical methods to assay enantiopure substances, and methods to determine configurations. Starting 'chiral pools' to create chiral products must be created. Molecular devices such as switches (including catenanes and rotaxanes as described on page 324 of "Emergent Computation: Emphasizing Bioinformatics") are described. Switches energized chemically or photo-driven. Molecular 360° rotary motors are described as are self-assembling nanotubes. Chiral Fullerenes composed of graphite may be formed into cages, nanotubes, nanocoils, nanocones, etc. Uses including artificial photosynthetics, and optically active helical polymers. DNA nanotechnology including knotted DNA molecules are described. Just as the Appendix in "Emergent Computation: Emphasizing Bioinformatics" discussed shape grammar applications of planar (and higher dimensional applications) of DNA, these are also discussed in this exciting paper.
"Light-Activated Hydrogel Formation via the Triggered Folding and Self-Assembly of a Designed Peptide", by L. A. Haines, K. Rajagopal, B. Ozbas, D. A. Salick, D. J. Pochan, J. P. Schneider, Journal of the American Chemical Society, Dec. 7 2005, 127, 48, 17025 - 17029
Hydrogels are dilute polymer networks capable of encapsulating large volumes of water. Applications include drug delivery and wound healing tissue engineeringm and are currently extensively used in the fabrication of contact lenses. In vivo applications must be cytocompatible but in many cases are in fact cytotoxic at low concentrations. This paper reports the use of a 20-residue photocaged peptide VKVKVKVKVDPPTKVKXKVKV–NH2 with hairpins that is unfolded in aqueous solution under ambient light, but under exposure to UV radiation, results in an uncaged peptide with intramolecular folds. This work has similarities to emergentcomputation.com/quad.html#PHOTO for recent work with quadruplexes.
"DNA duplex-quadruplex exchange as the basis for a nanomolecular machine", by P. Alberti, J.-L. Mergny, Proceedings of the National Academy of Sciences USA, Feb. 18 2003, 100, 4, 1569 - 1573
intramolecular quadruplex + C-fuel → linear dsDNA
linear dsDNA + G-fuel → intramolecular quadruplex + GC dsDNA waste
The above cycle results in a contraction between the quadruplex conformation and the linear conformation.
"A Single DNA Molecule Nanomotor", by J. J. Li, W. Tan, Nano Letters, April 1 2002, 2, 4, 315 - 318
A nanomotor is constructed based upon two conformations of DNA: intermolecular duplex (linear) DNA changing to obtain intramolecular tetraplex conformation (ω=TGGTTGGG, thus the tetraplex: ωTGTωR). The motor cycles ten times in 4000 seconds.
Back to TopNano Tiled DNA
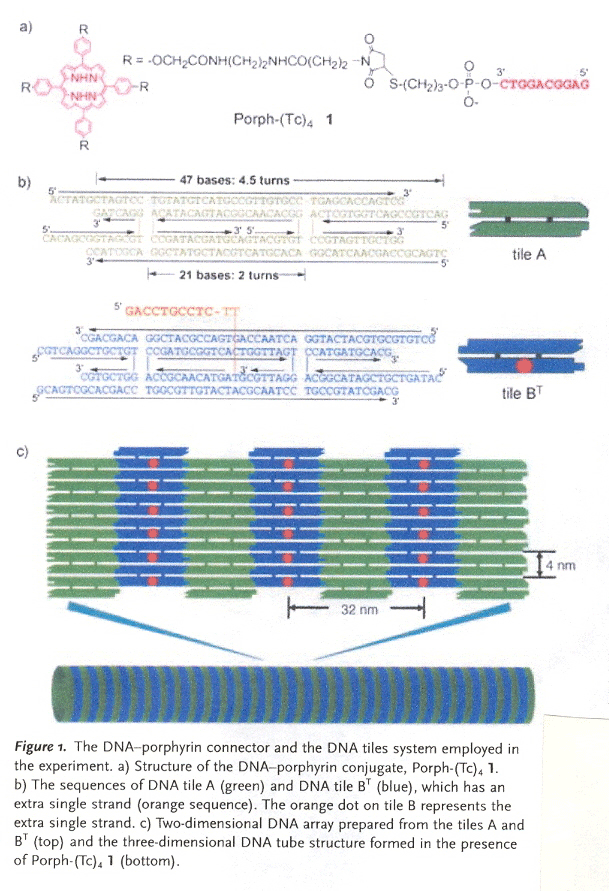
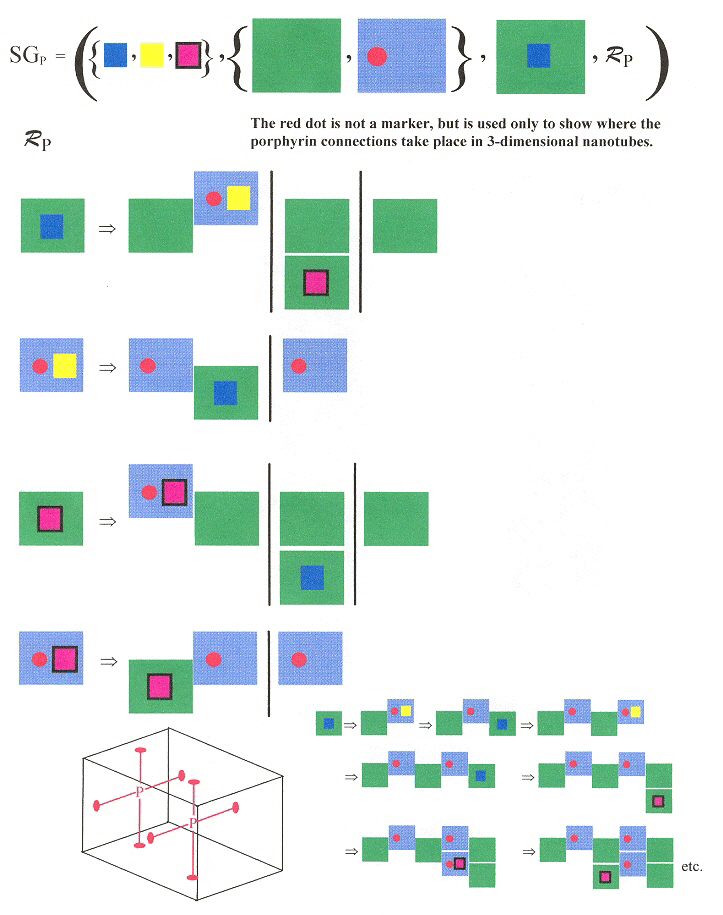
"DNA Tube Structures Controlled by a Four-Way-Branched DNA Connector", M. Endo, N. C. Seeman, T. Majima, Angewandte Chemie, 2005, 44, 6074 - 6077
A porphyrin connector composed of four identical groups "R". "R" has a Watson-Crick complementary tag strand "Tc" to a tag "T" on Tile BT. Without the porphyrin connector, tiles A and BT form a two-dimensional DNA array (thus tile BT has four different sticky-end overhangs plus the additional tag T). With the connector present, T and Tc self-assemble into 3-dimensional nanotubes. Tiles A and BT have been construced with Watson-Crick overhanging sticky-ends to induce the observed 2-dimensional self-assembly.
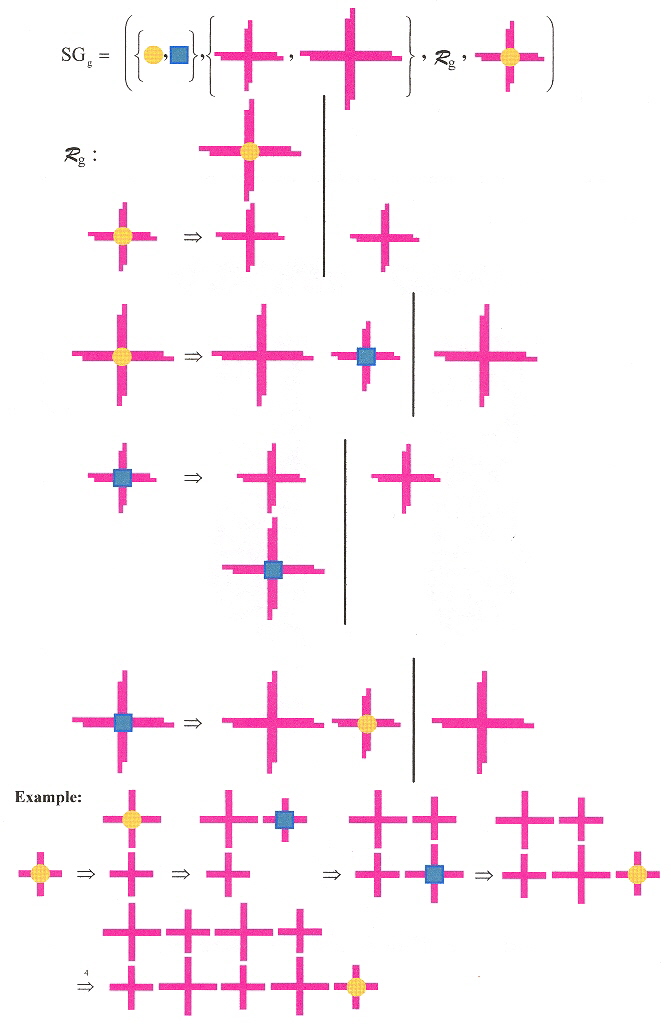
"DNA-Templated Self-Assembly of Two-Dimensional and Periodic Gold Nanoparticle Arrays", by J. Sharma, R. Chhabra, Y. Liu, Y. Ke, H. Yan, Angewandte Chemie International Edition, Jan. 23 2006, 45, 5, 730 - 735
Gold nanoparticles conjugated with thiolated DNa bind to DNA A tiles. These A tiles are then mixed with three different kinds of B tiles to obtain NGAB (Nano Gold AB), NGABL (Nano Gold AB Long), and NTAB (Nano gold Track AB) self-assembling structures. The DNA is in the form of a 2-dimensional Holliday structure (cruciform) as discussed in the appendix in "Emergent Computation: Emphasizing Bioinformatics". Some Holliday-like structures have longer arms (thus the Long structures). Some have only 3 sticky ends (hence the 1-dimensional Track structures). The researchers used novel methods to be able to obtain reliable results. The researchers state that by using variable sized Holliday-like structures, reliable and programmable spacing of gold nanoparticles in 1- and 2-dimensions can be achieved. The researchers note that the cruciform structures are not perfectly square, and that gold nanoparticles sometimes detach from the DNA. The researchers forsee new materils with different kinds of nanoparticles such as the following.
- Photonic antenna nanoparticles
- Magnetic nanoparticles
- Semiconductor nanoparticles
A shape grammar very similar to the kind discussed in the appendix of "Emergent Computation: Emphasizing Bioinformatics" can easily be constructed, where two classes of Holliday structures are used, the elements in each class being of different sizes.
Shape Grammar Sg
Nano Proteins
"Aptamer-Directed Self-Assembly of Protein Arrays on a DNA Nanostructure", by Y. Liu, C. Lin, H. Li, H. Yan, Angewandte Chemie International Edition, 2005, 44, 28, 4333 - 4338
DNA self-assembly begins with the chemical synthesis of single-stranded DNA that self-assemble into branched DNA known as 'tiles'. DNA tiles can carry sticky ends that are complementary to the sticky ends of other DNA tiles, thus resulting in the assembly of tiles into DNA lattices. One application of DNA-based self-assembly is the use of self-assembled DNA nanostructures to direct the assembly of other macromolecular components such as proteins. Aptamers are DNA or RNA molecules selected for their ability to bind to other molecules, such as other nucleic acids, proteins, organic compounds, and organisms. This paper reports selective DNA aptamer binding to link proteins to periodic sites of a self-assembled DNA array. Different tiles and lattices with different geometric patterns can be designed. A DNA grammar for such tiles induces a grammar on the induced protein lattice. As discussed in the appendix to "Emergent Computation: Emphasizing Bioinformatics", if a shape grammar is used to describe the DNA lattice, the induced grammar for the proteins would also be the corresponding shape grammar for the protein lattice.
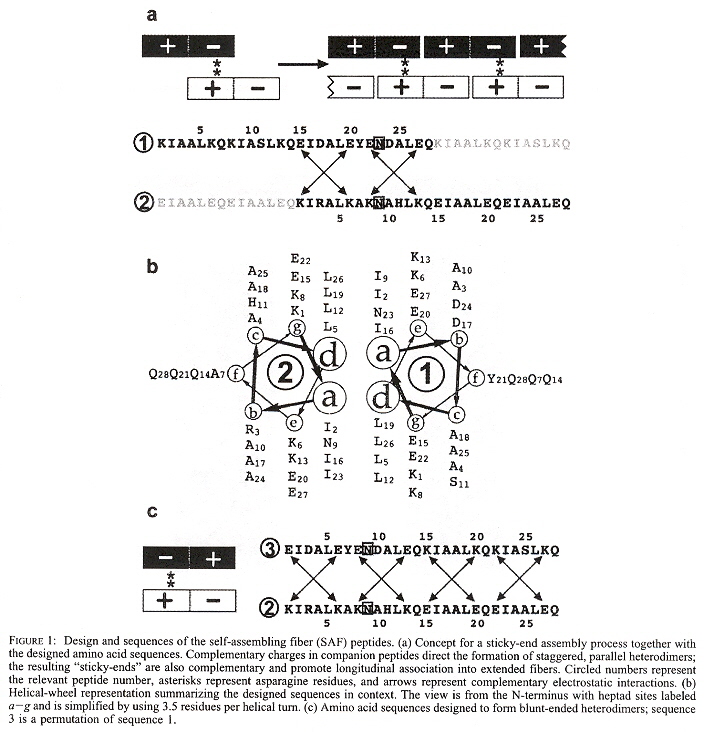
"Sticky-End Assembly of a Designed Peptide Fiber Provides Insight into Protein Fibrillogenesis", by M. J. Pandya, G. M. Spooner, M. Sunde, J. R. Thorpe, A. Rodger, D. N. Woolfson, Biochemistry, Aug. 1 2000, 39, 30, 8728 - 8734
Using H to represent a hydrophobic amino acid, and P to represent a polar amino acid,
then the heptad repeat is of the form (HPPHPPP)n. Two 28-residue peptides
have been designed to form a staggered (analogous to "sticky ends) heterodimer used for assembly of
"coiled-coil" fibers. SAF abbreviates
Each SAF polypeptide is a heptad of the form abcdefg as follows: isoleucines at a, leucines at d, glutamate at e, lysine at g, asparagine at a in complementary heptads. SAF-p3 designed as a circular permutation of SAF-p1, swapping the last two heptads with the first two heptads. SAF-p1 with SAF-p2 produces a fibre, SAF-p2 with SAF-p3 is a blunt dimer.
See the Figure below. "b" looks down from the top (N terminus) towards the bottom (C terminus), abcdefg on the left is circled as 2, as it is SAF-p2, while abcdefg on the right is circled as 1, as it is SAF-p1. The amino acid subscripts refer to the numberings in "a".
SAFs
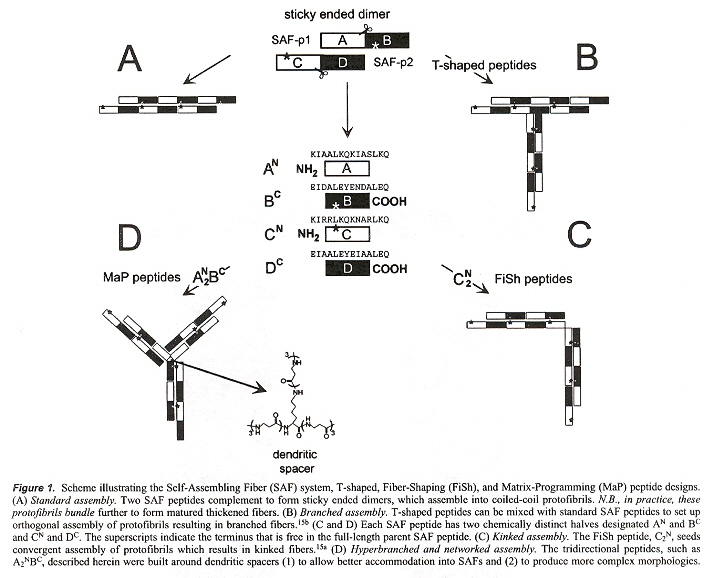
"MaP Peptides: Programming the Self-Assembly of Peptide-Based Mesoscopic Matrices", by M. G. Ryadnov, D. N. Woolfson, Journal of the American Chemical Society, Sept. 7 2005, 127, 35, 12407 - 12415
This paper is very similar to the 2000 paper. Each SAF polypeptide is a heptad of the form abcdefg as follows: Ile at a, Leu at d, Lys at e and g, at the N terminus. Glu for e and g at the C terminus to obtain staggered assembly. Asparagine at a different a site. in complementary heptads. SAF-p1 with SAF-p2a produces a fibre. AN BC couples with CN DC with DC associated with AN (see the figure below).
Staggered SAFs
"Wrapping Peptide Tubes: Merging Biological Self-Assembly and Polymer Synthesis", by M. A. B. Block, S. Hecht, Angewandte Chemie International Edition, Nov. 4 2005, 44, 43, 6986 - 6989
Membranes are importan to living things, and transmembrane channels and pores that combine biological self-assembly with integratwed polymer synthesis is the study in this paper. Various tubular structures composed of amino axcid building blocks are explored by different laboratories. For example, α-peptide strands with alternating D- and L-α-amino acids twisted to form a single-stranded β-helix. Other motifs are mentioned, but there are three major outstanding problems:
- Control of the length of self-assembled tubes.
- Controlled functionalization of both the interior and exterior surfaces of self-assembled tubes.
- The hierarchical organization of isolated tubular building blocks organized as hybrid materials.
Idealized Self-Assembling Peptide Tubular Structures
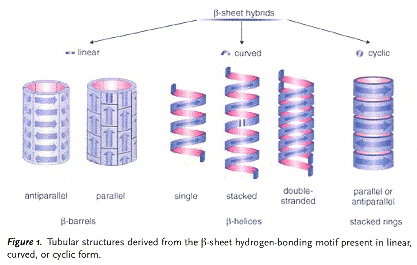
"Laminated Morphology of Nontwisting β–Sheet Fibrils Constructed via Peptide Self-Assembly", by M. S. Lamm, K. Rajagopal, J. P. Schneider, D. J. Pochan, Journal of the American Chemical Society, Nov. 30 2005, 127, 47, 16692 - 16700
Nearly all β–sheet fibrils reported in the literature thus far have an intrinsic helical pitch or twist. The synthetic materials reported in this paper are composed of a peptide sequence that exhibits a nontwisted morphology (under the conditions tested). The peptide sequences of interest are:
- (VK)4—V·[DPLP]·T—(KV)4
- (VK)4—V·[LPLP]·T—(KV)4
The first peptide sequence forms β–hairpin structures that assemble reversibly, the second peptide sequence assembles irreversibly into extended β–strands that are highly ordered two-dimensional nanostructures. Assembly is controlled by various stimuli such as pH, temperature, and ionic strength, and these stimuli are used to control reversibility.
Back to Topi-Motif Nanotechnology
"A Proton-Fuelled DNA Nanomachine", by D. Liu, S. Balasubramanian, Angewandte Chemie International Edition, 2003, 42, 46, 5734 - 5736
Nanomachines fuelled by compementary ddDNA oligonucleotides often accumulate dsDNA waste products that poison the system, decreasing efficiency, until they rapidly become useless. This paper reports a four-stranded i-motif "quadruplex" that is stable, based upon the following complementary strands.
X 5'–CCCTAACCCTAACCCTAACCC–3'
Y 3'–GATTGTGATTGTGATTG–5'
At pH 5.0, X folds into an i-motif structure. At pH 8.0, X unfolds and is hybridized with Y
to get a duplex dsDNA. Switching between conformations is rapid, the waste product dsDNA is
nontoxic. The paper reports 30 cycles over 5000 seconds controlled only by pH.
"DNA Molecular Motor Driven Micromechanical Cantilever", by W. Shu, D. Liu, M. Watari, C. K. Riener, T. Strunz, M. E. Welland, S. Balasubramanian, R. A. McKendry, Journal of the American Chemical Society, 2005, 127, 48, 17054 - 17060
This paper is a good application of the above paper ("A Proton-Fuelled DNA Nanomachine"). This
paper reports upon the integration of i-motif structure nanomachines with a mechanical
nanodevice (a cantilever), to show that a nanomachine can actually be assembled to do work. The
idea is to attach an array of dsDNA oligonucleotides with an alkanethiol linker to an array of
cantilevers. Using pH to control i-motif to dsDNA interconversion, the force exerted due to
conformational change bends the cantilever in a specific direction (to do work). The conformtional
changes are reversible allowing a cycle of cantilever bending, without poisoning the motor. The
authors point out that in contrast to polymers and rotaxanes, these motors operate under mild,
aqueous conditions, and selection of oligonucleotide sequences supports control of a tuneable
system. The oligonucleotide sequences reported in this are as follows (the underlined base
mismatches in Y are to prevent intramolecular G-quadruplex formation.
X HSC6H12–5'–CCC AAT CCC AAT CCC AAT CCC–3'
Y 3'–GTG ATT GGG ATT TGG ATT GTG–5'
"Duplex Pinching: A Structural Switch Suitable for Contractile DNA Nanoconstructions", by R. P. Fahlman, M. Hsing, C. S. Sporer-Tuhten, D. Sen, Nano Letters, Aug. 2003, 3, 8, 1073 - 1078
G-quartets in an intramolecular structure can "pinch" or contract, acting as a nanoswitch. The "pinching" or contracting is easily controlled by modifying cation concentration by using a chelator. The two states of contraction are linear dsDNA vs a DNA G-quartet, as depicted in quad.html#QQQQ2.
Back to TopPhilosphy of Nanotechnology
"Inventing the Nanomolecular Wheel", by J. Siegal, Science, Oct. 7 2005, 310, 5745, 63 - 64
A number of papers have been published establishing the latest conquests in designing and constructing a variety of nanotechnological objects. Objects such as nanotubes, nanosprings, nanogears, nano-propellers, etc. It would appear that the approach taken is based upon a form of philosophical "reductionism". How long before nano-cars, nano-ships, nano-lawnmowers, etc.? Thus this article discusses the report in Science about a nano-rotory-motora. Not to disparage the hopes that nanotechnology might provide for mankind, but a closer examination of some of the philosophical questions is a worthy effort. The question is asked "Why are some scientists so quick to believe in such direct parallels between macro- and nanomachinery?" Siegel asks "What are the metaphorical infidelities of the classical molecular model, and what is their impact on Fletcher et al.'s molecular rotor and the dream of nanomachines?"
"Despite the time-averaged correlated motion of these gears, their motion is essentially Brownian and without the capacity to do work." Siegel points out that over multiple cycles, these motors do work by "using enthalpy to corral entropy, something clearly different from their macroscale counterparts." Thus the macroscale machines do work by using thermal fluctuations, while Brownian machines at the nano-level have rapidly dissipated thermal gradients and losses of energy due to collisions with solvent molecules. The point being the following: "The appeal of direct molecular analogs to macroscopic machines makes paying attention to the distinctions between macro- and nanoscopic mechanisms all the more crucial."
a "A Reversible, Unidirectional Molecular Rotory Motor Driven by Chemical Energy", by S. P. Fletcher, F. Dumur, M. M. Pollard, B. L. Feringa, Science, Oct. 7 2005, 310, 5745, 80 - 82
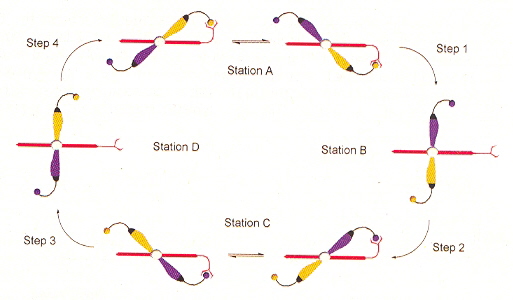
"DNA-programmed assembly of nanostructures", by K. V. Gothelf, T. H. LaBean, Organic Biomolecular Chemistry, 2005, 3, 4023 - 4037
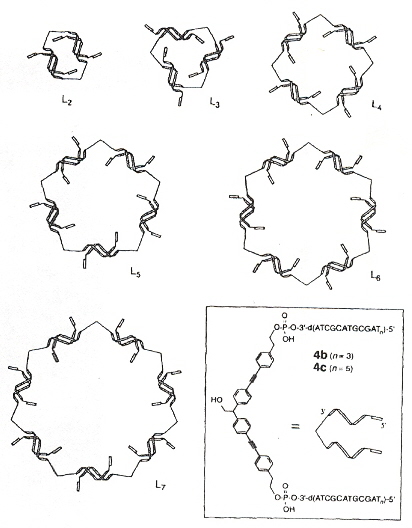
A good review of the general trends in DNA self-assembly of nanostructures. Discussion of the use of synthetic oligonucleotides as well as other molecules such as PNA. Two and three dimensional nanostructures are discussed, using modules composed of multiple building blocks. The living cell is viewed as the most advanced nanostructure based upon self-assembly of different building blocks. Given a choice between DNA, RNA and proteins, proteins are considered the lesser choice due to their higher complexity. Double-helix tiles are discussed, but triple-helix DNA is also discussed a. Organic self-assembling cyclic scaffolds such as L2, L3, through L7 are discussed b (see the figure, below). Many other experimental results are discussed in this paper.
Self-Assembly of Cyclic DNA