A disclaimer: finding scientific articles dealing with the subjects covered in "Emergent Computation: Emphasizing Bioinformatics" requires a great deal of work and time. The major reason for this is that the subjects of Bioinformatics from the point of view of Mathematical Linguistics as well as applications of Mathematical Linguistics in areas such as Biology, Meteorology, Oceanography, Geology, Chemistry, etc. are not yet recognized as a discipline of study. As a consequence, it is not claimed that the scientific articles or books cited here constitute all the relevant articles that may have been published, just those that have come to light.
Other References to G-quartets/Quadruplexes
Mitochondrial (mtDNA), Chloroplast (ctDNA), kinetoplast (kDNA), Plasmid DNA, Virus DNA & Viroid RNA)
Quadruplex structures are very significant. One major area of significance is that quadruplex stability differes from the Watson-Crick model. In the Watson-Crick model, A/T (U) and C/G form stable complements. Hoogsteen bonds stabilize X-quadruplexes where X can be A, C, G, T, and iso-G, etc. Thus, G-G, C-C, A-A, T-T structures are stable. This is a very significant departure from the usual Watson-Crick motif.
Many new and very exciting things will be reported here. One major thing to be noted is that one of the major mathematical methods of examining DNA from the point of view of linguistics will be found to be quite limited, limited in large part as a consequence of ignoring or underrating biochemistry. Here we will see that Splicing Systems and Dominoes are quite limited. Some of these limitations include the following:
Index
Terminolgy: planar Tetrad vs 3D Quadruplex

"Electrochemical Telomerase Assay with Ferrocenylnapththalene Diimide as a Tetraplex DNA-Specific Binder", by S. Sato, H. Kondo, T. Nojima, S. Takenaka, Analytical Chemistry, Nov. 15 2005, 77, 22, 7304 - 7309
While this paper is what it purports to be (an assay to detect tetraplexes), it also illustrates in a very practical way, how tetraplexes actually might arise. In this assay, ferrocenylnapththalene diimide is used to bind to DNA in tetraplex form. In the first image, below, we see (in schematic or cartoon fashion), the tetraplex folding induced by telomerase. In the second figure, we see how through simple "winding" of a single strand of DNA (or RNA) can form a tetraplex.
Telomerase action
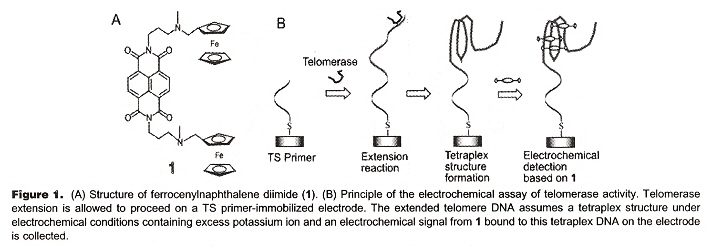
|
"TS" abbreviates Telomerase Substrate
|
Inter- and Intramolecular Quadruplex Formation
|
"Characterization of the Structural Conformation Adopted by (TTAGGG)n Telomeric DNA Repeats of Different Length in Closed Circular DNA", by D. Huertas, H. Lipps, F. Azorín, Journal of Biomolecular Structure & Dynamics, Aug. 1994, 12, 1, 79 - 90
G-quadruplexes are reported within circular plasmid DNA. Specifically, the plasmids pHU3, pHU6, pHU8, pHU13, pHU18, and pHU21 are examined, where pHUn plasmid contains (TTAGGG)n.
"G-QUARTET STRUCTURES IN TELOMERIC DNA", by J. R. Williamson, Annual Review of Biophysics and Biomolecular Structure, 1994, 23, 703 - 730
This paper is a wonderful resource of information! Among other things, different syn/anti conformations in quadruplex structures are clearly shown. Multiple kinds of polymorphism are explained. Cleavage of quadruplexes by Topoisomerase II is also discussed.
"Eukaryotic topoisomerase II cleavage of parallel stranded DNA tetraplexes", by I. K. Chung, V. B. Mehta, J. R. Spitzner, M. T. Muller, Nucleic Acids Research, April 25 1992, 20, 8, 1973 - 1977
While it is known that triplex DNA blocks topoisomerase II cleavage, and does not cleave single stranded DNA, yet
topoisomerase II is reported here to cleave parallel tetraplex DNA. Specifically, ssDNA oligo M as tetraplex
M4 is cleaved at two sites in the oligonucleotide, below. M (an ssDNA) is not cleaved however.
oligo M is the 47-base oligonucleotide:
5'–TAGTCCAGGCTGAGC ↓ AGGT ↓ ACGGGGGAGCTGGGGTAGATGGGAATGT–3'
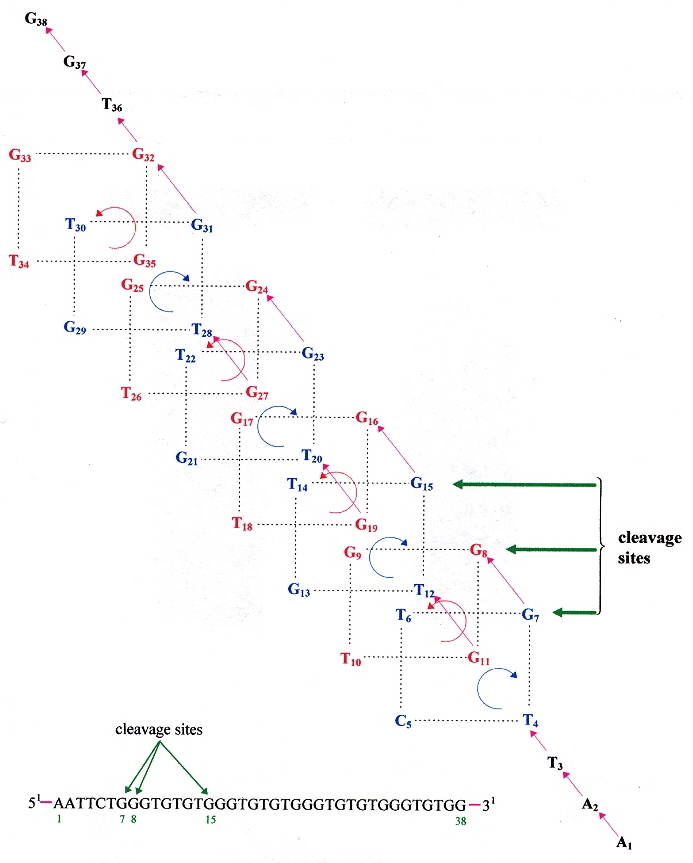
"Saccharomyces cerevisiae Mre11 is a high-affinity G4 DNA-binding protein and a G-rich DNA-specific endonuclease: implications for replication of telomeric DNA", by G. Ghosal, K. Muniyappa, Nucleic Acids Research, 2005, 33, 15, 4692 - 4703
Saccharomyces cerevisiae protein ScMre11p not only binds to G-rich G4 ssDNA, but cleaves telomeric quartets as an endonuclease. The cleavages follow:
G-Quartet Endonuclease Cleavage
"Formation of a PNA2—DNA2 Hybrid Quadruplex", by B. Datta, C. Schmitt, B. A. Armitage, Journal of the American Chemical Society, April 9 2003, 125, 14, 4111 - 4118
FRET (flourescence resonance energy transfer) experiments to study the effects of PNA2—DNA2 in d(G4T4G4) G-quadruplexes. The following paper discusses PNA/DNA hybrids at length, but this paper discusses the use of FRET techniques used in studying these hybrids.
|
PNA/DNA Flourophore Structures
|
PNA/DNA FRET Combinations
|
"Topology Variation and Loop Structural Homology in Crystal and Simulated Structures of a Bimolecular DNA Quadruplex", by Hazel, P., Parkinson, G. N., Neidle, S., Journal of the American Chemical Society, April 26 2006, 128, 16, 5480 - 5487
The specific topology of DNA quadruplexes is investigated. Specifically, the following molecules are investigated:
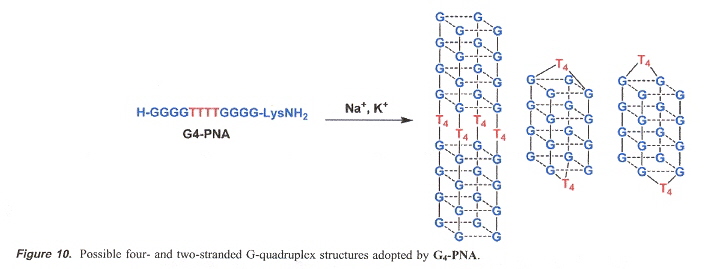
"Quadruplex Formation by a Guanine-Rich PNA Oligomer", by B. Datta, M. E. Bier, S. Roy, B. A. Armitage, Journal of the American Chemical Society, March 30 2005, 127, 12, 4199 - 4207
A Peptide Nucleic Acid with the structure H-G4T4G4-Lys-NH2 forms a four-stranded quadruplex.
PNA Quadruplex Oligomer
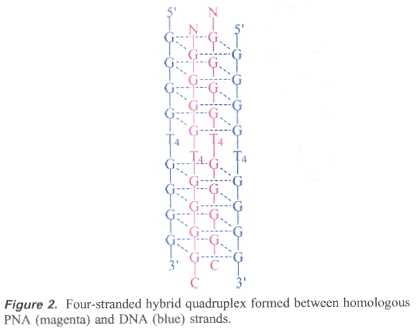
This was suggested by the already-known quadruplex formed by a hybrid PNA with a DNA (of homologous strucrture).
PNA/DNA Quadruplex
It is thus possible then that more complicated PNA quadruplexes might exist.
G-PNA Quadruplex
"RNA Guanine Quadruplex Invasion by Complementary and Homologous PNA Probes", by V. L. Martin, B. A. Armitage, Journal of the American Chemical Society, June 8 2005, 127, 22, 8032 - 8033
RNA aptamer RDQ is hybridized with its corresponding complementary PNA to form a G-quadruplex. Experimental results depend upon the fact that the G-quadruplex is hypochromic, while the duplex structure is hyperchromic. PNA probes P7C complements the blue region in Chart 1, as does P7CTO (which is fluorescent when in a quadruplex structure). Experiment shows that the both PNA probes P7C and P7CTO hybridize with the blur region of RDQ. PNA probes P7H and P7HTO are both fluoescent, but now with stoichiometry of 2:1 (P7C/P7CTO have stoichiometry 1:1). Thus when P7H and P7HTO hybridize, they invade the quadruplex (see the figure to the right).
|
RDQ Quadruplex with PNA Probes
|
RNA Quadruplex-PNA Invasion
|
"Hybridization of Complementary and Homologous Peptide Nucleic Acid Oligmers to a Guanine Quadruplex-Forming RNA", by V. L. Marin, B. A. Armitage, Biochemistry, 2006, 45, 1745 - 1754
Guanine-rich PNAs can form hybrid PNA2–RNA quadruplexes. P7C hybridizes with the blue strand in the "RNA Quadruplex" in the above right figure.
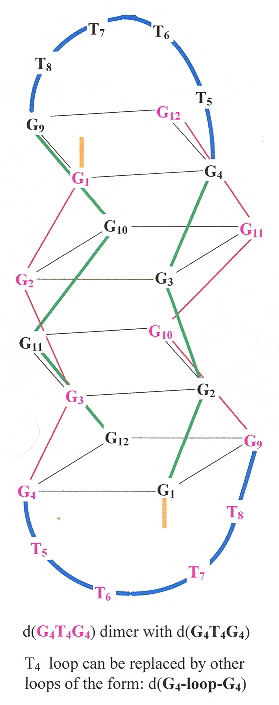
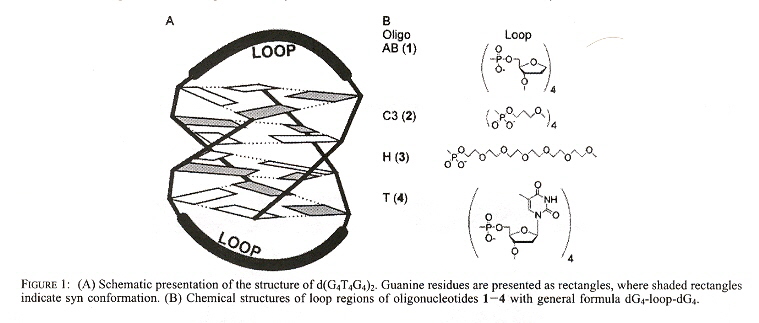
Back (to TOP of page)"Role of Loop Residues and Cations on the Formation and Stability of Dimeric DNA G-Quadruplexes", by M. Cevec, J. Plavec, Biochemistry, Nov. 22 2005, 44, 46, 15238 - 15246
G-quadruplexes of dimeric sequences of the form dG4-loop-dG4 are examined, under the influence of Na+, K+, and NH4+. The first Figure shows the G-quadruplex, the following Figure shows the G-quadruplex anti-syn arrangement along with the different "loop" chemical structures.
G-quadruplex for dG4-T4-dG4
G-Quadruplex Anti-Syn Arrangement and Different "loops"
"A Dimeric RNA Quadruplex Architecture Comprised of Two G:G(:A):G:G(:A) Hexads, G:G:G:G Tetrads and UUUU Loops", by H. Liu, A. Matsugami, M. Katahira, S. Uesugi, Journal of Molecular Biology, Oct. 4 2002, 322, 5, 955 - 970
RNA also folds into a quadruplex template containing a G-Tetrad mixed with a G:A-hexad.
RNA G-Quartet Self-Assembly
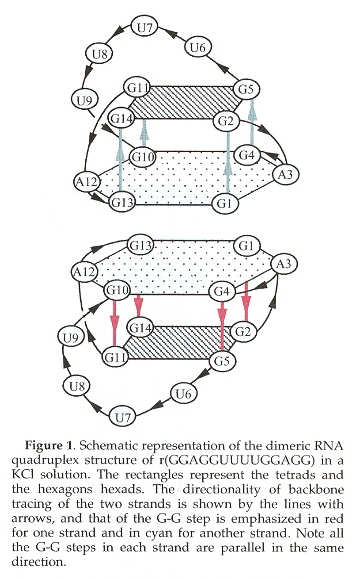
"A DNA pentaplex incorporating nucleobase quintets", by J. C. Chaput, C. Switzer, Proceedings of the National Academy of Sciences U.S., Sept. 14 1999, 96, 19, 10614 - 10619
Nucleobases can be designed for a DNA pentaplex. Quadruplexes generally require the presence of K+, but pentaplexes requires CS+ and are constructed with iG (2'-deoxy-iso-guanosine) as [T4iG4]5.
DNA Pentaplex Layer
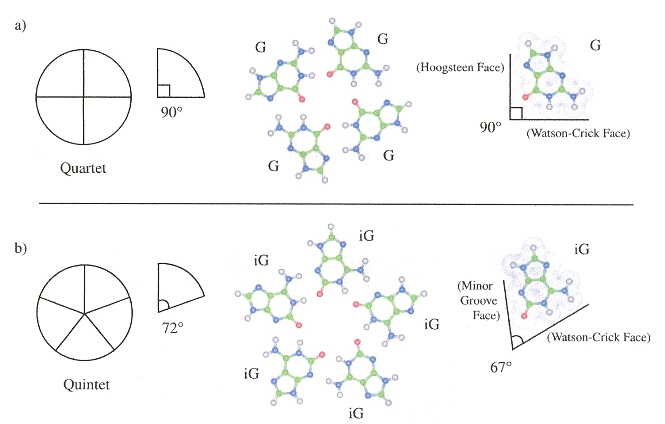
DNA Pentaplex
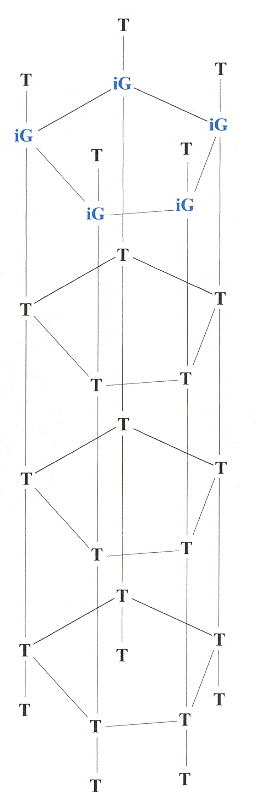
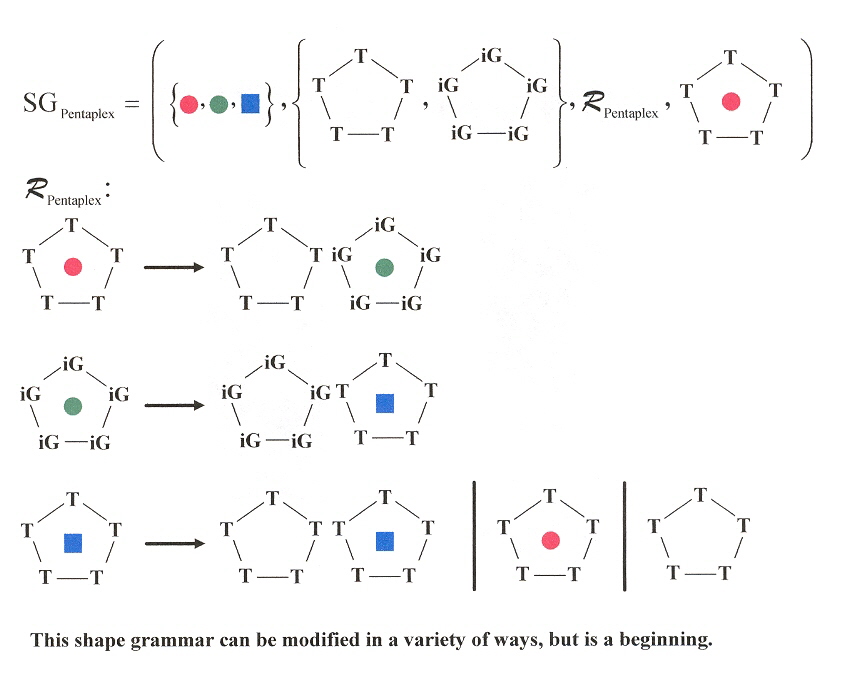
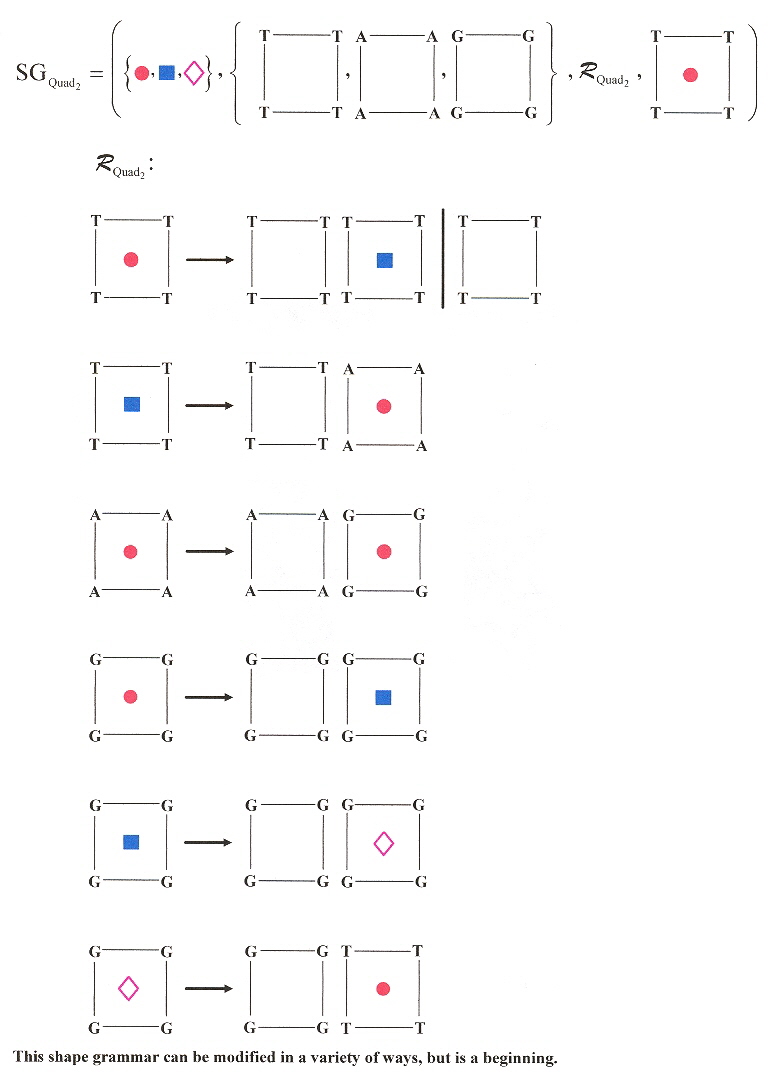
In the following 3-dimensional Shape grammar for a similar Pentaplex language, imagine each plane progressively stacked above the following plane.
DNA Pentaplex 3-D Shape Grammar
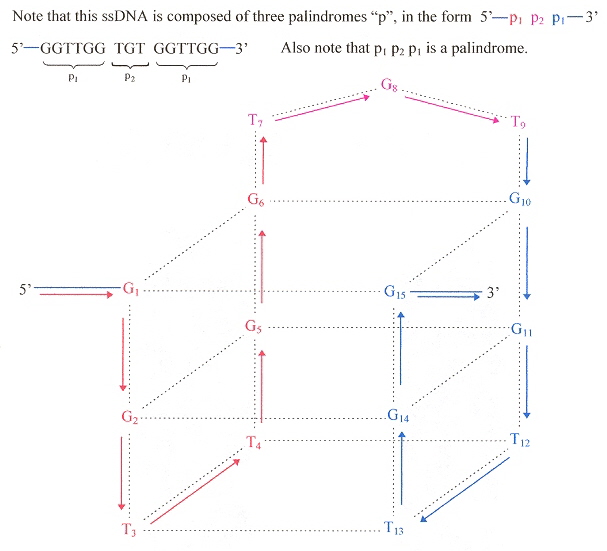
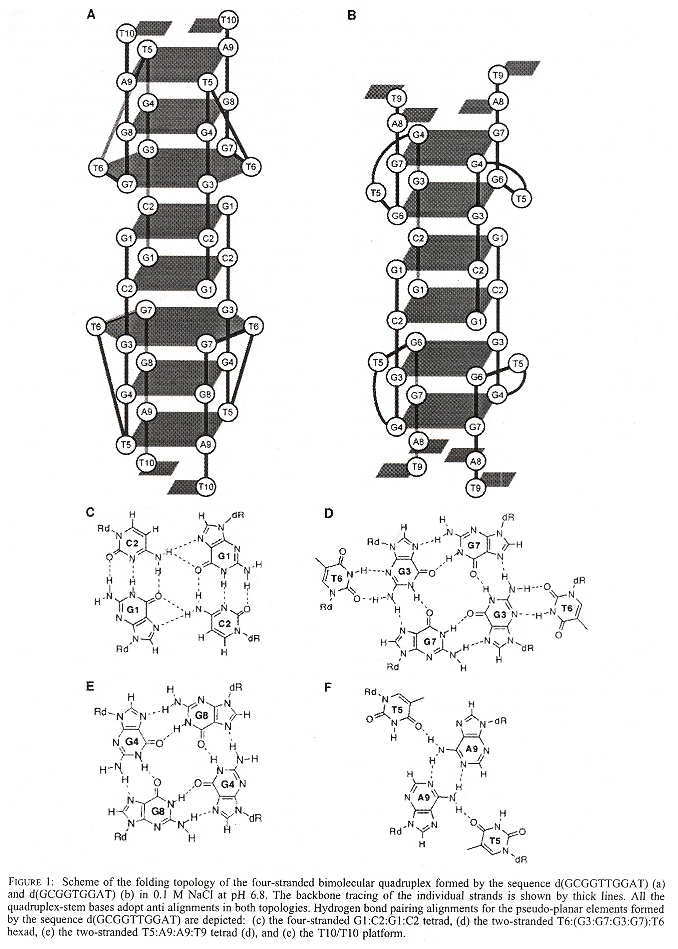
"Experimental Demonstration of T:(G:G:G:G):T Hexad and T:A:A:T Tetrad Alignments within a DNA Quadruplex Stem", by M. W. da Silva, Biochemistry, March 15 2005, 44, 10, 3754 - 3764
Using the special sequence d(GCGGTTGGAT), a quadruplex mixed with a hexaplex or heptaplex results that is pH-sensitive. Other special sequences used included: d(GCGGUTGGAT) and d(GCGGTUGGAT). Note the GGTTGG palindrome, as well as the "almost" palindromes GGUTGG, and GGTUGG.
Mixed Quadruplex/Hexaplex
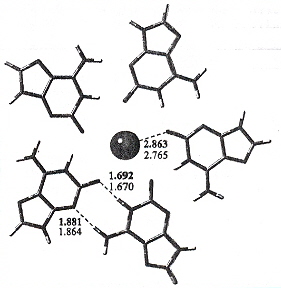
"Isoguanine Complexes: Quintet versus Tetrad", by J. Gu, J. Leszczynski, Journal of Physical Chemistry B, 2003, 107, 6609 - 6613
Isoguanine (isoG) forms stable quintets, especially in the presence of K+. The bonds are between
isoG Quintets
"Studies on the synthesis of a G-rich octaoligoisonucleotide (isoT)2(isoG)4(isoT)2 by the phosphotriester approach and its formation of G-quartet structure", by J. Chen, L. R. Zhang, J. M. Min, L. H. Zhang, Nucleic Acids Research, 2002, 30, 13, 3005 - 3014
Parallel G-quadruplex structures that incorporate isoT and isoG are observed.
Back (to TOP of page)"Intercalative G-Tetraplex Stabilization of Telomeric DNA by a Cationic Porphyrin", by I. Haq, J. O. Trent, B. Z. Chowdhry, T. C. Jenkins, Journal of the American Chemical Society, 1999, 121, 12, 1768 - 1779
Three different kinds of quadruplexes can be constructed by using three different oligonucleotide strings:
Quadruplexes
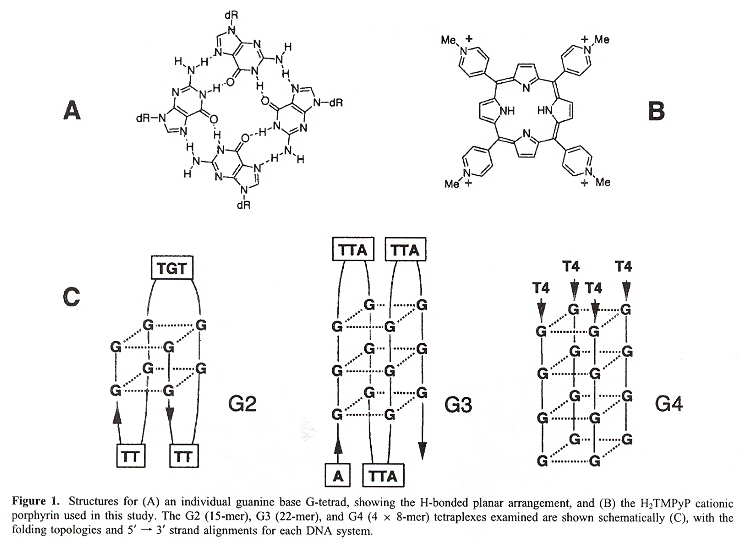
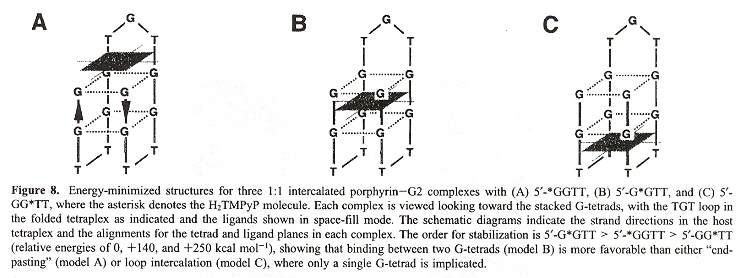
In the figure above, the G2 type quadruplex is found in a thrombin-binding aptamer telomere; the G3 type quadruplex is found in human single-stranded telomeres; the G4 type quadruplex is found in Oxytrichia telomeres. The point of this paper is that considering that porphyrin is approximately the same size and shape as the G-tetrad, possibly the porphyrin could act as a stabilizing molecule. In fact, the authors do find that porphyrin does act to stabilize the G2 type of quartet (below).
Porphyrin Stabilized Quadruplex (Chair Form)
In the following 3-dimensional Shape grammar for a similar Quadruplex language, imagine each plane progressively stacked above the following plane.
DNA Quadruplex 3-D Shape Grammar 1
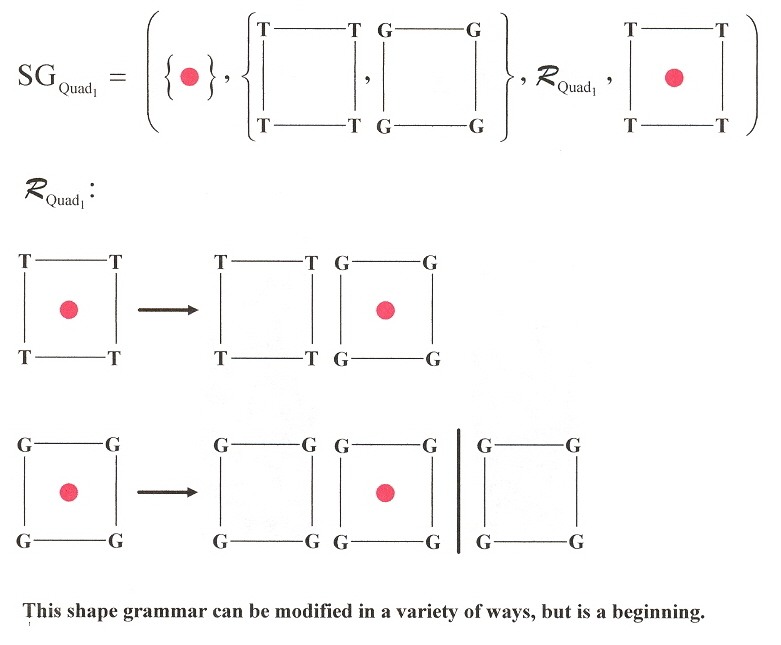
Another similar paper dealing with tetrads and porphyrins is the following paper. This paper discusses TMPyP4 or 5,10,15,20-tetra-(N-methyl-4-pyridyl)porphine and TMPyP2 or 5,10,15,20-tetra-(N-methyl-2-pyridyl)porphine. The paper also shows how 5'–CATG GTGG TTT (GGGTTA)4 CCAC–3' can fold into an intramolecular hairpin, then refold into different G-quadruplex conformations, as well as three-layered and six-layered dimer guadruplexes stabilized by TMPyP4.
"Interactions of TMPyP4 and TMPyP2 with Quadruplex DNA. Structural Basis for the Differential Effects on telomerase Inhibition", by F. X. Han, R. T. Wheelhouse, L. H. Hurley, Journal of the American Chemical Society, April 21 1999, 121, 15, 3561 - 3570
3- & 6-layered quadruplex dimers with Porphyrin stabilizers
Porphyrin stabilizers are the black squares
"Stabilization of guanine quadruplex DNA by the binding of porphyrins with cationic side arms", by T. Yamashita, T. Uno, Y. Ishikawa, Bioorganic & Medicinal Chemistry, April 1 2005, 13, 7, 2423 - 2430
d(TTAGGG) antiparallel quadruplex conformation is used to study the effects of peripheral positional isomers of porphyrin. To be explicit, the effects of steric hinderance at the para vs meta positions of the peripheral tolyl groups. The results show that there are two possible stable ways that such porphyrin groups may bind to a quadruplex: either external-stacking, or by groove-binding mode.
|
Porphyrins ("p" for para, "m" for meta)
|
External-Stacking vs Groove-Binding
|
"Formation of a Complex of 5, 10, 15, 20 –Tetrakis(N–methylpyridinium–4–yl)–21H,23H –porphyrin with G–Quadruplex DNA", by H. Mita, T. Ohyama, Y. Tanaka, Y. Yamamoto, Biochemistry, June 6 2006, 45, 22, 6765 - 6772
TmPyP4 intercalates between G–C base pairs as well to A–T base pairs, as well as to duplex RNA and DNA—RNA hybrids, and to triplex DNA.
"A Spectroscopic Study on the Interactions of Porphyrin with G-Quadruplex DNAs", by C. Wei, G. Jia, J. Yuan, Z. Feng, C. Li, Biochemistry, May 30 2006, 45, 21, 6681 - 6691
Porphyrinoids are discussed in their role of stabilizing G-Quadruplexes not only in positions peripheral to terminal tetrads in G-Quadruplexes, but also intercallated at non-peripheral positions between tetrads in G-Quadruplexes.
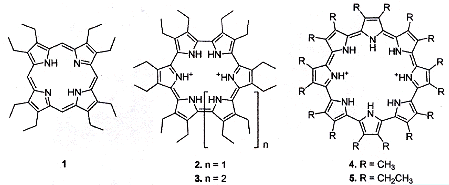
"Cyclo[n]pyrroles: Size and Site-Specific Binding to G-Quadruplexes", by E. S. Baker, J. T. Lee, J. L. Sessler, M. T. Bowers, Journal of the American Chemical Society, March 1 2006, 128, 8, 2641 - 2648
This interesting paper discusses "porphyrinoids" in G-quadruplexes for
T4 = d(TTAGGG)4. Various molecules are discussed, but central to the discussion are
1 = octaethylporphyrin,
2 = cyclo[6]pyrrole,
3 = cyclo[7]pyrrole,
4 and 5 = cyclo[8]pyrrole (see below).
Porphyrinoids
T4 = d(TTAGGG)4 can have multiple conformations, with porphyrinoids (see below).
T4 Porphyrinoid Conformations
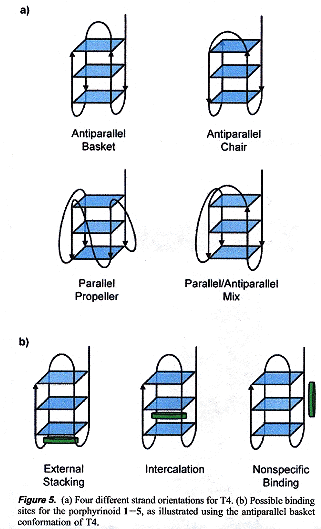
Results found are that the binding strengths are 2 > 1 > 3 > 4 >> 5, while size is 1 < 2 < 3 < 4 < 5. Size and charge (only 1 = octaethylporphrin has a neutral charge) are found to determine structure and stability. 2 = cyclo[6]pyrrole is most stable as this molecule binds most strongly to the G-quadruplex.
"Light-Induced Formation of G-Quadruplex DNA Secondary Structures", by G. Mayer, L. Kröck, V. Mikat, M. Engeser, A. Heckel, ChemBioChem, Nov. 2005, 6, 11, 1966 - 1970
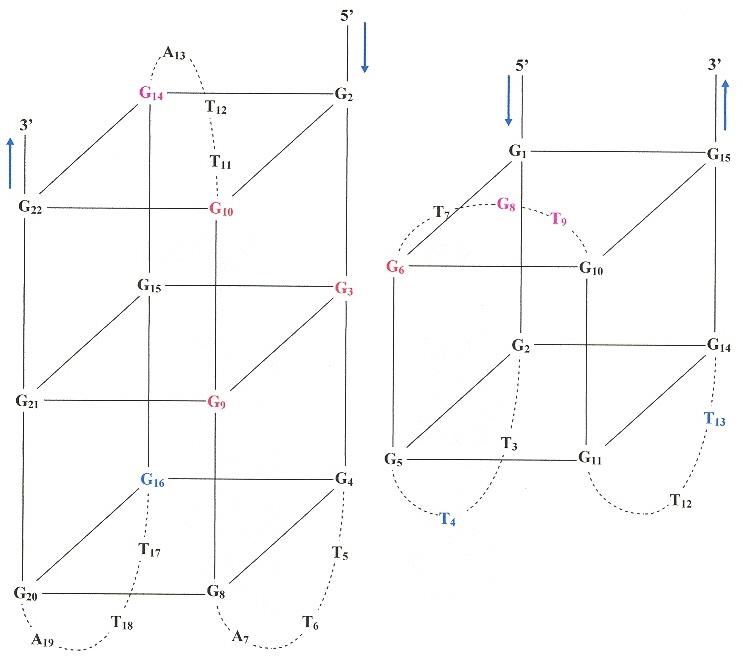
A "caged" molecule has protecting groups that mask their biological activity (are sterically constrained). In this paper, "caging" is photolabile and can be deactivated by irradiation by light, the caging being accomplished by using phosphoramidite (see the first figure, below), where dGNPP is used. Previous papers a, b discussed the use of caged dTNPP. Thus by irradiation, it is possible to remove the caged protection, to permit dynamic quadruplex formation. Specific positions in an oligonucleotide sequence are more important in qudruplex formation. Examining the second figure, below oligonucleotide 7 forms a qudruplex, but oligonucleotide 8 does not form a quadruplex until it is irradiated. Similarly, the oligonucleotide 12 does not form a quadruplex until it is irradiated (both examples have dG located in the sensitive central G-plane). Bases in other positions are less significant to quadruplex folding (9, 10, and 11). Similarly, oligonucleotide 13 with only two layers forms a quadruplex, as does 14 once it is irradiated, but the less significant positions in oligonucleotides 15 through 18. do not affect quadruplex formation upon irradiation.
dGNPP and Oligonucleotides
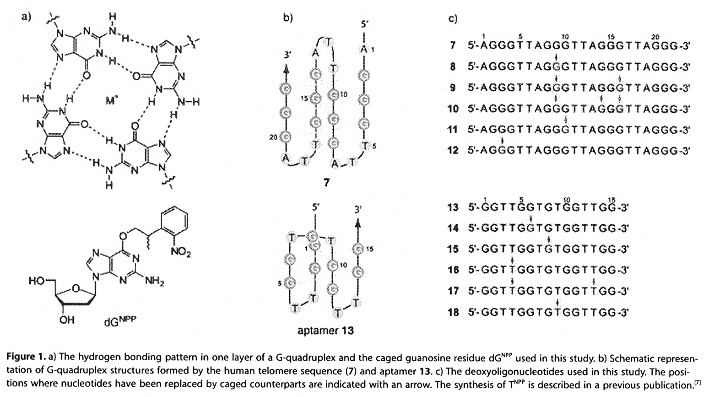
Photolabile Caged Quadruplexes
a "Light regulation of aptamer activity: an anti-thrombin aptamer with caged thymidine nucleobases", by A. Heckel, G. Mayer, Journal of the American Chemical Socoety, Han. 26 2005, 127, 3, 822 - 823
b "Photo induced Transcription by Using Temporarily Mismatched Caged Oligonucleotides", by L. Kröck, A. Heckel, Angewandte Chemie International Edition, 2005, 44, 3, 471 - 473
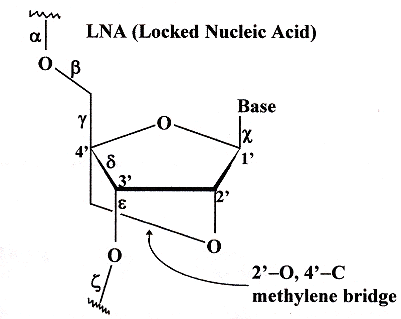
"NMR solution structures of LNA (locked nucleic acid) modified quadruplexes", by J. T. Nielsen, K. Arar, M. Petersen, Nucleic Acids Research, April 13 2006, 34, 7, 2006 - 2014
Examining the figure below, we note the 2'–O to 4'–C methylene bridge. This bridge reduces conformational possibilities, hence such sugar–nucleotide bases are referred to as LNA (locked nucleic acid) bases. LNA have been found to confer stability upon dsDNA, triple-helix DNA, as well as in G–Quadruplexes. Thus G–Quadruplexes composed of d(TG4)T) have less stability than their corresponding d(TL4)T) and d(TGLGLT) analogues where "L" refers to any LNA.
Locked Nucleic Acid
Similarly, ENA also functions in the same manner as LNA, where ENA has a 2'–O to 4'–C ethylene bridge (see below) a
ENA
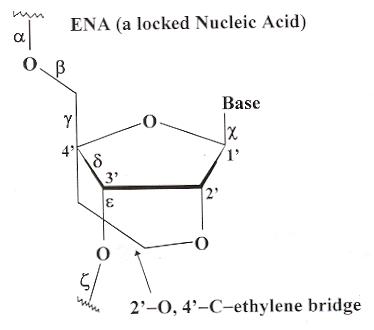
a "Conformationally restricted triplex-forming oligonucleotides (TFOs). Binding properties of α-L-LNA and introduction of the N7-glycosylated LNA-guanosine", by A. A. Koskin, Tetrahedron, June 19 2006, 62, 25, 5962 - 5972
"Spectroscopic studies of the interaction between Quercetin and G-quadruplex DNA", by H. Sun, Y. Tang, J. Xiang, G. Xu, Y. Zhang, H. Zhang, L. Xu, Bioorganic & Medicinal Chemistry Letters, July 1 2006, 16, 13, 3586 - 3589
G-quadruplexes are of medical/pharmaceutical value. To have medical and pharmaceutical value, G-quadruplex stability must be increased, and a number of classes ofsmall molecules confer stability, but are often toxic. Quercetin in a small molecule that is a plant flavonoid and has no toxic side effects but confers G-quadruplex stability. Quercitin binds at terminal ends of G-quadruplex structures, but also has a second binding mode. Monomeric binding is intercalative, but when binding dimeric G-quadruplexes, Quercetin binds along the DNA groove.
Back (to TOP of page)"Synthesis and Evaluation of Quindoline Derrivatives as G-Quadruplex Inducing and Stabilizing Ligands and Potential Inhibitors of Telomerase", by J-L. Zhou, Y-J. Lu, T-M. Ou, J-M. Zhou, Z-S. Huang, X-F. Zhu, C-J. Du, X-Z. Bu, L. Ma, L-Q. Gu, Y-M. Li, A. S-C. Chan, Journal of Medicinal Chemistry, Nov. 17 2005, 48, 23, 7315 - 7321
Telomerase adds telomeric repeated sequences onto the ends of a telomere. In this paper, some derivatives of quindoline interact with the G-quadruplex d[G3(T2AG3) 3] in preference to duplex DNA, with the following properties:
|
Quindoline
|
Quindoline Derivatives
|
"Fluorescent Amplifying Recognition for DNA G-Quadruplex Folding with Cationic Conjugated polymer: A Platform for Homogeneous Potassium Detection", by F. He, Y. Tang, S. Wang, Y. Li, D. Zhu, Journal of the American Chemical Society, Sept. 7 2005, 127, 35, 12343 - 12346
The essential idea of this paper is that single-stranded DNA has a much longer linear distance than when it folds (under the influence of K+) into G-quartets with a much smaller and compact distance. As fluoresence depends greatly upon Förster distance (1/r6), the G-quartet form will fluoresce much more.
"G-Quartets 40 Years Later: 5'-GMP to Molecular Biology and Supramolecular Chemistry", by J. T. Davis, Angewandte Chimie International Edition, Jan. 30 2004, 43, 6, 668 - 698
A review of G-quartets, with heavy emphasis upon self-assembly, useful in nanotechnology.
"Structure of hnRNP D Complexed with Single-stranded Telomere DNA and Unfolding of the Quadruplex by Heterogeneous Nuclear Ribonucleoprotein D", by Y. Enokizono, Y. Konishi, K. Nagata, K. Ouhashi, S. Uesugi, F. Ishikawa, M. Katahira, The Journal of Biological Chemistry, May 13 2005, 280, 19, 18862 - 18870
Heterogeneous nuclear riboprotein D or hnRNP (also known as AUF1) binds specifically to ssDNA sequence d(TTAGGG)n, r(UUAGGG)n in mRNA. This paper reports that hnRNP inhibits the formation of RNA quadruplex.
"Natural and synthetic G-quadruplex interactive berberine derivatives", by M. Franceschin, L. Rossetti, A. D'Ambrosio, S. Schirripa, A. Bianco, G. Ortaggi, M. Savino, C. Schultes, S. Neidle, Bioorganic & Medicinal Chemistry Letters, 2006, 16, 1707 - 1711
As with porphyrins and other molecules such as acridines, triazines, perylenes, and anthraquinones, berberine may be associated with G-quartets for reasons of stability, pharmaceutical properties, inhibition of telomerase, etc. berberine is an antibiotic with anti-cancer activity, it inhibits telomere elongation, and binds to G-quadruplexes. This paper discusses some of these G-quadruplex binding properties.
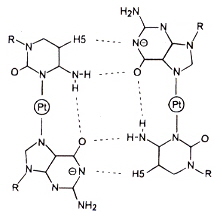
Back (to TOP of page)"A Metalated Guanine, Cytosine Base Quartet with a Novel GC Pairing Pattern Involving H(5) of C", by S. Metzger, B. Lippert, Journal of the American Chemical Society, Dec. 11 1996, 118, 49, 12467 - 12468
Metal ions are found to be capable of stabilization in place of hydrogen-bonding in G2C2-base quartets. It would take little work to create a shape grammar for such quartets.
A G2C2-base Quartet Stabilized by Metal Ions in place of H-bonding
"Structure of the parallel-stranded DNA quadruplex d(TTAGGGT)4 containing the human telomeric repeat: evidence for A-tetrad formation from NMR and molecular dynamics simulations", by E. Gavathiotis, M. S. Searle, Organic & Biomolecular Chemistry, May 21 2003, 1, 10, 1650 - 1656
Hoogsteen bonds as opposed to Watson-Crick complements yield A-quartets as well as the well known G-quartets.
|
G and A Quartets
|
A-Tetrad based upon d(TTAGGGT)4
|
"Synthesis and Characterization of DNA Quadruplexes Containing T-Tetrads Formed by Bunch-Oligonucleotides", by G. Oliviero, J. Amato, N. Borbone, A. Galeone, M. Varra, G. Piccialli, L. Mayol, Biopolymers, Feb. 15 2006, 81, 13, 194 - 201
This paper adds to information about A-tetrads, T-tetrads, C-tetrads, G-tetrads, and mixed tetrads, as well as tetrads of modified bases. This paper is based upon bunch tetrads such as bunch-[d(TG4T)]4 in which the four 3'-ends are linked together by a tetrabranched spacer. Two specific molecules are of interest in this paper: bunch-[d(TGGTGGC)]4 and bunch-[d(CGGTGGT)]4. T-tetrads are found to be mixed with G-tetrads.
"NMR Observation of a Novel C-Tetrad in the Structure of the SV40 Repeat Sequence GGGCGG", by P. K. Patel, N. S. Bhavesh, R. V. Hosur, Biochemical and Biophysical Research Communications, April 21 2000, 270, 3, 967 - 971
A C-tetrad composed of four d-TGGGCGGT sequences that stack over a G4-tetrad (but poorly over a G6-tetrad).
C-Tetrad
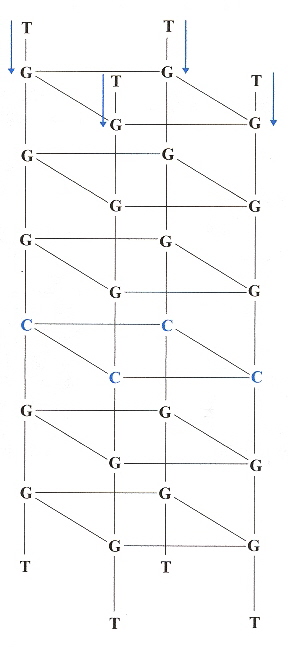
"Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter, by A. T. Phan, V. Kuryavyi, H. Y. Gaw, D. J. Patel, Nature Chemical Biology, Aug. 2005, 1, 3, 167 - 173
Most G-quadruplex structures consist of four-guanine-tract sequences. In addition, some experiments included inosine. This paper also includes studies concerning the stability of different ligands with one of the G-qudruplexes studied, specifically Pu24I (the abbreviation from Purine-rich strand with 24 nucleotide bases, including the base inosine). Ligands included the following.
The molecular structures of the first three ligands follow. For porphyrin, See this reference.
Ligands
Some of the Pu24I strands of interest, included the following:
Examining the figure below in the case of Pu24I, there are three G-tetrads:
Five-Guanine-tract G-quadruplex
The ligands stacked above the top G-tetrad (G4•G8•G13•G17).
In the following 3-dimensional Shape grammar for a similar Quadruplex language, imagine each plane progressively stacked above the following plane.
DNA Quadruplex 3-D Shape Grammar 2
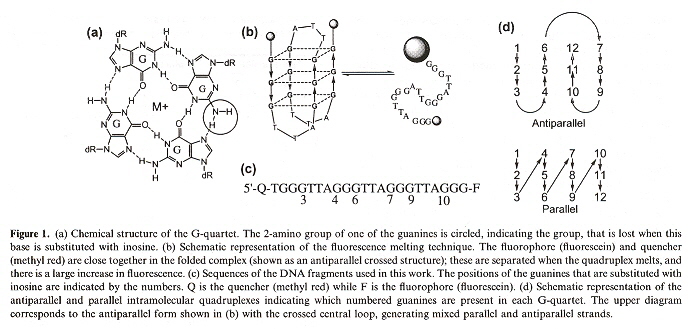
"Inosine substitutions demonstrate that intramolecular DNA quadruplexes adopt different conformations in the presence of sodium and potassium", by A. Risitano, K. R. Fox, Bioorganic & Medicinal Chemistry Letters, April 15 2005, 15, 8, 2047 - 2050
This paper studies guadruplexes of [(GGGTTA)3GGG] in which one of the guanines is substituted with inosine. The data for potassium suggests a parallel arrangement of the strands, while for sodium there is a mix of both parallel and antiparallel forms. A single inosine replaces guanine at positions 3, 4, 6, 7, 9 and 10 to determine structure. Different folding results in a fluoresence signal indicated by Flouroscene Resonance Energy Transfer (FRET).
Inosine Parallel & antiparallel Quadruplexes
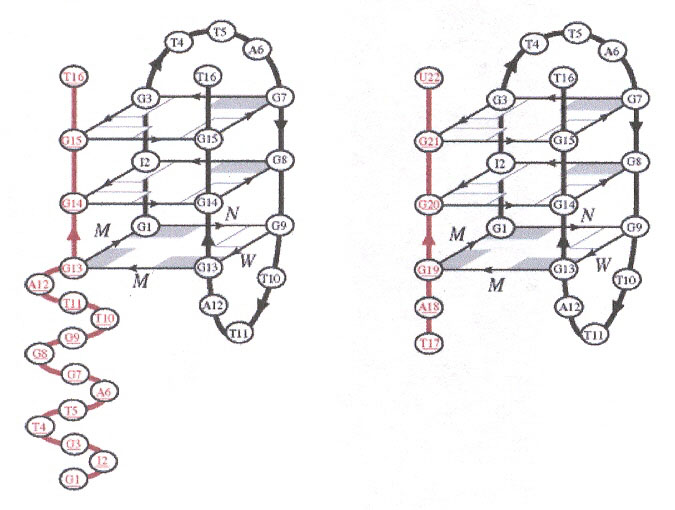
"(3+1) Assembly of Three Human Telomeric Repeats into an Asymmetric Dimeric G-Quadruplex", by N. Zhang, A. T. Phan, D. J. Patel, Journal of the American Chemical Society, Dec. 14 2005, 127, 49, 17277 - 17285
There are two significant results reported in this paper. First, two strands of human telomeric d(GIGTTAGGGTTAGGGT) associate to form a three G-repeat telomeric sequence, as opposed to the usual one-, two-, or four-repeat sequences. Second, the G-quadruplex can form from two strands of greatly different length [d(TAGGU) with d(GIGTTAGGGTTAGGGT)]. Note that the strands can contain Inosine and the nucleotide bases T and U, as well.
2 × d(GIGTTAGGGTTAGGGT) and d(GIGTTAGGGTTAGGGT) with d(TAGGGU)
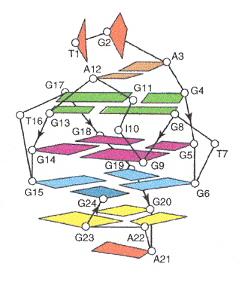
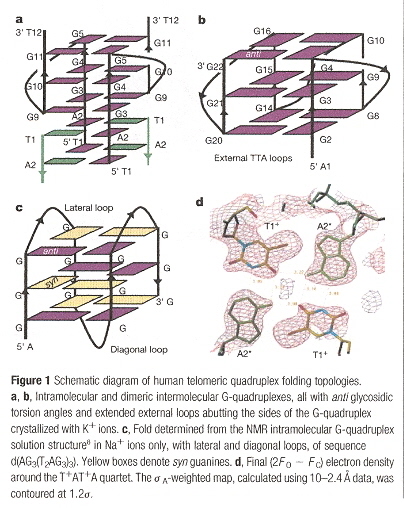
"Crystal structure of parallel quadruplexes from human telomeric DNA", by G. N. Parkinson, M. P. H. Lee, S. Neidle, Nature, June 20 2002, 417, 6891, 876 - 880
This paper very clearly discusses the "propeller" structured quadruplex, the anti and syn structural arrangements found in quadruplexes, and lateral and diagonal external loops. Rather than being overly wordy, the figures are extremely helpful.
In Figure 1, we see in a external loops from G5 to G9. In b, we see other external loops from G4 to G8, and G16 to G20. in c, lateral and diagonal loops are clearly identified. Furthermore, in c, the anti vs syn arrangements are clearly labelled as well.
External, Lateral, Diagonal Loops, Anti vs Syn Conformations
With G in green; T & U in blue; A in red, we can easily see the TTA "propeller" conformation in the next three figures, below.
|
TTA External Loop forming the 'Propeller'
|
TTA External Loop forming the 'Propeller'
|
TTA External Loop forming the 'Propeller'
|
"Solution Structure of an Unusually Stable RNA Tetraplex Containing G- and U-Quartet Structures", by C. Cheong, P. B. Moore, Biochemistry, 1992, 31, 8406 - 8414
r(UG4U)4 forms a stable parallel stranded quartet. Normal Watson-Crick or Hoogsteen bonds are replaced by Hydrogen bonds from the Nitrogen at position 3 with Oxygen from the Carbon at position 4. Thus there are four planes composed of G-quartets, samndwiched bwtween outer planes of Uracil-quartets.
r(UG4U)4 Parallel U-Quartet
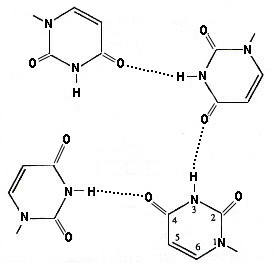
"Inverting the Charges of Natural Nucleobase Quartets: A Planar Platinum–Purine Quartet with Pronounced Sulfate Affinity", by M. Roitzsch, B. Lippert, Angewandte Chemie International Edition, Dec. 16 2005, 45, 1, 147 - 150
A synthetic G-quartet-like molecule is reported in which there is a restructuring of charges. Asside from synthetic quartets, synthetic triplets are also possible.
|
Synthetic Purine Quartet
|
Synthetic Purine–metal Architectures
|
"Intramolecular folding of a fragment of the cytosine-rich strand of telomeric DNA into an i-motif", by J.-L. Leroy, M. Guéron, J.-L. Mergny, C. Hélène, Nucleac Acids Research, May 1994, 22, 9, 1600 - 1609
"Stability of the I-motif Structure Is Related to the Interactions between Phosphodiester Backbones", by T. E. Malliavin, J. Gau, K. Snoussi, J.-L. Leroy, Biophysical Journal, June 2003, 84, 6, 3838 - 3847
"i-motif solution structure and dynamics of the d(AACCCC) and d(CCCCAA) tetrahymena telomeric repeats", by N. Esmaili, J.-L. Leroy, Nucleaic Acids Research, 2005, 33, 1, 213 - 224
"An unusual DNA structure detected in a telomeric sequence under superhelical stress and at low pH", by V. I. Lyamichev, S. M. Mirkin, O. N. Danilevskaya, O. N. Voloshin, S. V. Balatskaya, V. N. Dobrynin, S. A. Filippov, M. D. Frank-Kamenetskii, Nature, June 22 1989, 339, 626, 634 - 637
The 1989 paper immediately above points out that C-rich palindrome hairpins are paired with G-quadruplexe palindrome hairpins. This is a very important finding.
Some Quadruplex & i-motif possibilities
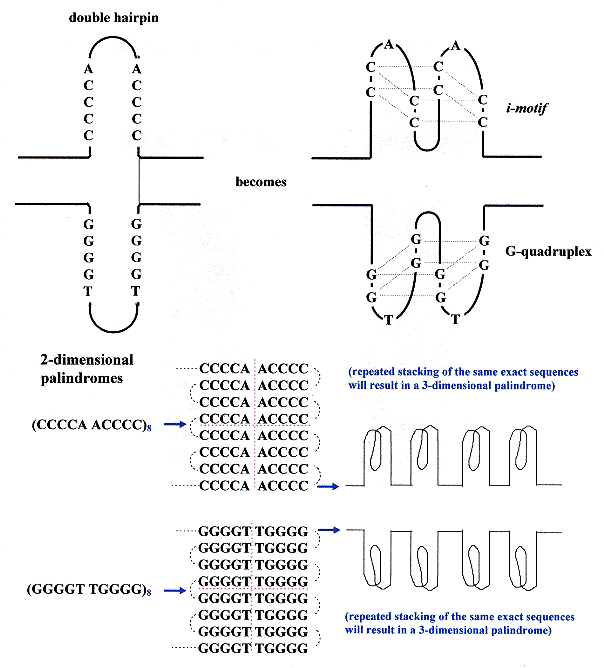
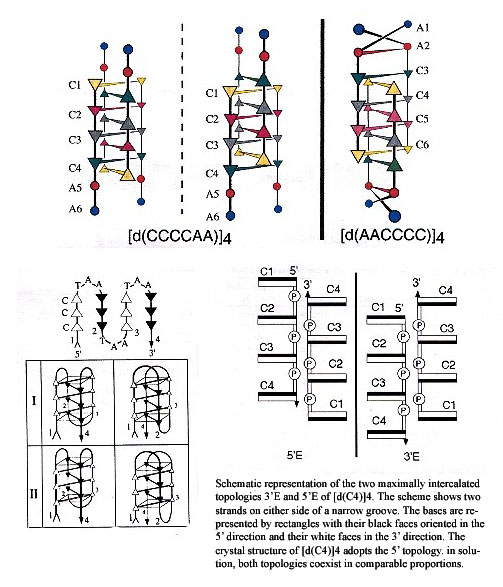
Two main types of tetrameric structures are known: G-quartets and the i-motif structure. The i-motif is built by the intercalation of two parallel duplexes, forming a rectangular-shaped structure, in which the positions of the four phosphodiester backbones define a wide and a narrow groove. It is the intercalation that is critical here. However, there exists two topologies for the i-motif structure — one with outermost 3' extremeties and the other with outermost 5' extremities, called, respectively, 3'E and 5'E topologies. Experimental investigation and simulation reveal that for [d(CC)]4, the 3'E topology has greater stability than the 5'E topology and indeed, the 5'E topology has not been observed experimentally. G-quadruplex and i-motif may be formed by association of four strands, by two hairpins containing two G- or C-rich stretches or by a folded single strand containing four G or C stretches. A possibility exists for the interaction of [d(CCCCAA)]4 and [d(AACCCC)]4 with the G-quartet of d(TTGGGG).
Intercalated Topologies of i-motif DNA
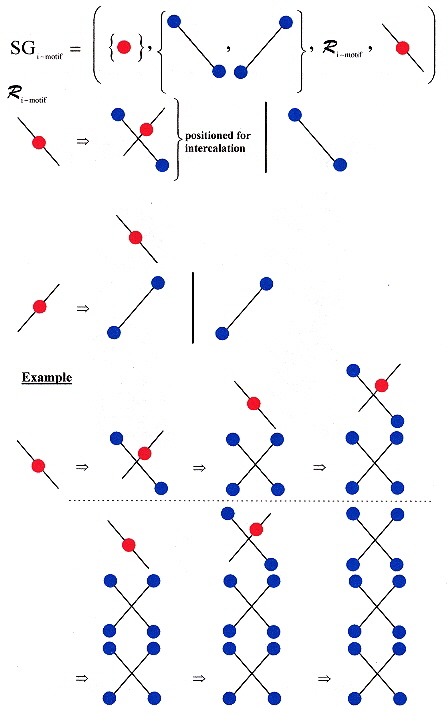
Shape Grammar for i-motif (intercalated) DNA
"Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb)", by Y. Xu, H. Sugiyama, Nucleic Acids Research, Feb. 7, 2006, 34, 3, 949 - 954
While G-quadruplex structures can form, associated complimentary sequences of C-rich DNA strands which are hemiprotonated
C-C+-base pairs adopt an i-motif structure. I-motif structures have been proposed to form in both centromeric
and telomeric regions of human chromosomes, as well as in the promoter region of the human c-myc gene.
Quadruplec-to-duplex conversion occurs more slowly at lower pH, while it is believed that low pH also stabilizes i-motif
structures. It is suggested that competing G-quadruplex and i-motif structure formation takes place at low pH
in vitro.
5'–GCCGCCCAAAACCCCCCG–3' from the 5' has many C bases,
forms the i-motif intercalated structure.
3'–CGGCGGGTTTTGGGGGGC–5' from the 5' has many G bases,
folds into a G-quadruplex.
"Deconvoluting the Structural and Drug-Recognition Complexity of the G-Quadruplex-Forming Region Upstream of the bcl-2 P1 Promoter", by Dexheimer, T. S., Sun, D., Hurley, L. H., Journal of the American Chemical Society, April 26 2006, 128, 16, 5404 - 5415
The expression of the P1 promoter of the human bcl-2 gene (proto-oncogenic, associated with many forms of cancer), is due to G-quadruplex formation. To be specific, the G-quadruplexes formed do not result in cell proliferation, but rather by reducing the rate of cell death. If expression (but not overexpression) of these G-quadruplexes can be enhanced, useful medical results might be possible. Such a program would only be useful if G-quadruplexes can be specifically targetted, not interfearing with other G-quadruplex activity. One way to approach this desired end is by targeting specific G-quadruplex by binding small molecules such as TMPyP4, Telomestatin, and Se2SAP.
"Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription", by Siddiqui-Jain, A., Grand, C. L., Bearss, D. J., Hurley, L. H., Proceedings of the National Academy of Science USA, Sept. 3 2002, 99, 18, 11593 - 11598
c-MYC oncogene expression can cause cancer-cell proliferation at Element lll1 (upstream from the P1 promoter). Element lll1 has an associated chair-form G-quadruplex that results from the folding-over of a DNA G-hairpin (as well as i-motif structures), and a single point mutation, G → A results in greatly increased c-MYC promoter transcription, thus c-MYC is a target for anti-cancer therapeutics. TMPyP4, by binding to G-quadruplex structures results in in vivo antitumor activity due to photoinduced DNA strand cleavage of external bridging loops (both to chair-form and basket-form quadruplexes). In some cases, insulin might also be treated by the same mechanism. Thus the mechanism is to bind TMPyP4 to a G-quadruplex, then using photoinduced cleavage to to destroy the G-quadruplex, therby destroying c-MYC oncogene expression.
"Cationic Porphyrins Promote the Formation of i-Motif DNA and Bind Peripherally by a Nonintercalative Mechanism", by O. Y. Fedoroff, A. Rangan, V. V. Chemeris, L. H. Hurley, Biochemistry, Dec. 12 2000, 39, 49, 15083 - 15090
It is reported that TMPyP4 or 5,10,15,20-tetra-(N-methyl-4-pyridyl)-porphine, promotes the formation of i-motif DNA structures (binds to and stabilizes G-quadruplex and i-motif structures). Most importantly, "Formation of a G-quadruplex structure within genomic DNA should therefore be coupled with the self-organization of the complementary C-rich strand". The authors state that "...i-motif interactions with either proteins or low-molecular weight ligands have not appeared, and to our knowledge, we present here the first such study. [d(AACCCC)]4 and its porphyrin complex are studied, as well as other C-rich telomeric repeats.
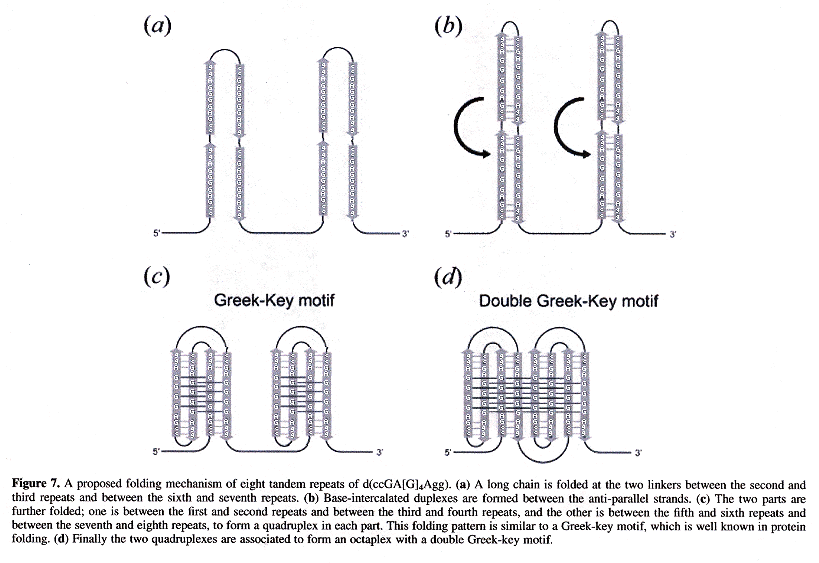
Back (to TOP of page)"Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggests a folding mechanism of eight tandem repeats", by J. Kondo, W. Adachi, S. Umeda, T. Sunami, A. Takénaka, Nucleic Acids Research, May 7 2004, 32, 8, 2541 - 2549
d(ccGA[G]4Agg) is of specific interest, where lower case base abbreviations refer to Watson-Crick bound base complements, the bases abbreviated in upper case participating in Hoogsteen quartet planes to form quadruplexes. Octaplex structures form where two intercalted I-motif G-quartets exist, looking like the figure below.
Intercalation of Octaplex I-motif Structures
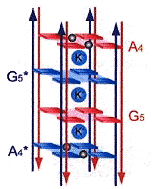
Using eight repeats of d(ccGA[G]4Agg) in the figure below, a ssDNA strand can form into a Double Greek-Key motif in which an octaplex with two intercalted G-quadruplexes exist! Other similar structures also form using d(tgGA[G]3Aca).
Double Greek-Key Octaplex I-motif Structures
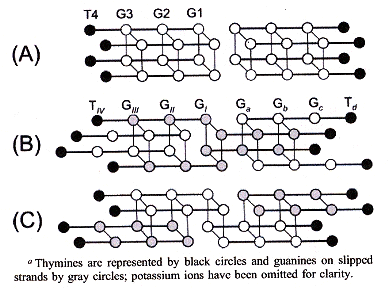
"Formation of an Interlocked Quadruplex Dimer by d(GGGT)", by Y. Krishnan-Ghosh, D. Liu, S. Balasubramanian, Journal of the American Chemical Society, Sept. 8 2004, 126, 35, 11009 - 11016
Molecule G3T = d(GGGT) forms two kinds of quadruplexes, named Ql and Qh. These two kinds of quadruplexes are distinguished by two –dA/dt vs Temperature peaks (at Tm = 38.5±1°C and Tm = 79±1°C, respectively). Both Ql and Qh form parallel quadruplexes. Ql is tetrameric [G3T]4, while Qh is octameric [G3T]8. Three possible octameric structures are possible ("Blunt-End", and "Interlocking" C2v, and C2h). While (G3T)4 is a parallel quadruplex, (G3T)8 is of the C2v-type interlocking structure (see below).
Possible (G3T)8 Structures
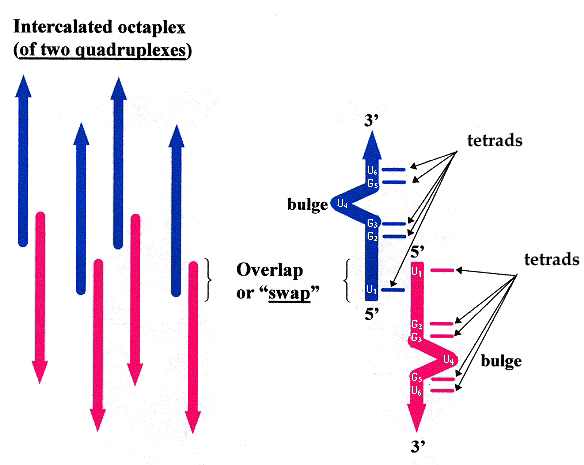
"Base-tetrad swapping results in dimerization of RNA quadruplexes: Implications for formation of the I-motif RNA octaplex", by B. Pan, K. Shi, M. Sundaralingam, Proceedings of the National Academy of Science USA, Feb. 28 2006, 103, 9, 3130 - 3134
In the hexamer 5'–U1-G2-G3-U4-G5-U6–3', base U4 forms a bulge, while the other nucleotide bases form tetrads. In addition, base U1 swaps positions with U1> in the opposing quadruplexes (see the figure below).
Quadruplex base U bulges and Swapping
"A comparative binding study of BIV Tat peptide against its TAR RNA duplex, RNA-DNA heteroduplex and DNA duplex", by J. B.-H. Tok, L. Bi, Bioorganic & Medicinal Chemistry Letters, 2005, 15, 129 - 133
As the 17-mer BIV Tat peptide has been observed to fold into a β-hairpin structure upon binding to its 26-mer TAR RNA target, this study addresses whether the corresponding RNA-DNA heteroduplexes and DNA duplex of TAR RNA retain Tat peptide binding. The result is that the RNA-DNA duplexes do bind, but not the DNA duplex.
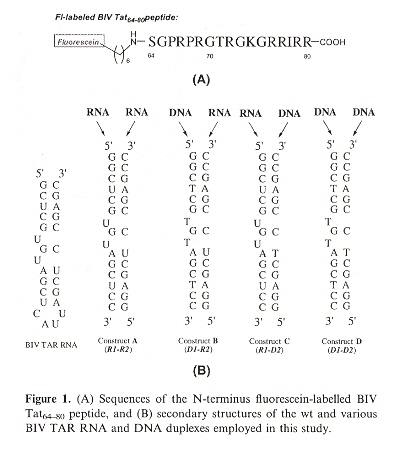
"Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis", by M. Nowotny, S. A. Gaidamakov, R. J. Crouch, W. Yang, Cell, July 1, 2005, 121, 1005 - 1016
Further confirmation that double stranded hybrids, one strand composed of RNA the other strand composed of DNA, are of significance. One area of significance concerns restriction enzymes, another area of significance concerns the adequacy of linguistic methods discussed in "Emergent Computation: Emphasizing Bioinformatics" ( p. 72 and p. 308). For example, Splicing systems do not cover such DNA/RNA hybrids. The few papers discussing DNA/RNA hybrids did not support detailed linguistic anaysis.
Generally, a triplex consists of a duplex where nucleotide bases are formed by Watson-Crick hydrogen bonds, and a third strand in which hydrogen bonds to the duplex are Hoogsteen bonds. Due to structural distortion of the sugar-phosphate backbone, the precondition of triplex formation is a homopyrimidine (Py) sequence in one duplex strand (with its complementary homopurine "Pu" sequence in the complementary strand). Thus duplex tracts of PyPu are required. The third strand in a triplex may be either Py or Pu. As a consequence, two types of triplexes form: PyPuPy (sometimes called 'H form') and PyPuPu. A preceeding superscript asterisk may be used to denote the third strand-base in a triple-base triad. Thus motifs such as CG·*C, TA·*T, TA·*A. Triplexes may form by intermolecular (one strand of the duplex folds over) or intramolecular (a single strand folds over twice) structures. Triads are as follows (first two are Watson-crick complements) a:
Circular Oligonucleotide Triple-Helix
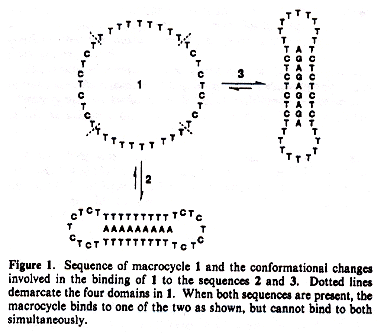
DNA/RNA heteroduplex hybrids exist, and linguistic methods must be sufficiently
comprehensive to include such structures. This discussion targets triplexes.
Triplexes are central to this discussion, as DNA/RNA hybrid triplexes also exist
(thus DNA/RNA hybrid triplexes also extend subject of linguistic analysis (as do
quadruplexes, pentaplexes, hexaplexes, and octaplexes).
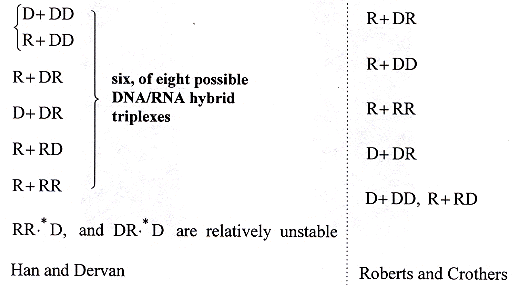
DNA/RNA hybrid triplexes may exist in eight varieties c. Notationally,
"D" will denote a strand of DNA, and "R" will denote a strand of RNA, then the
following types of DNA/RNA hybrid triplexes may be distinguished (order is top to
bottom, stability highest at the top). It should be noted that DNA/RNA hybrids
are targets for enzymes including ribonuclease H and reverse transcriptase
d.
The Six Stable RNA/DNA Hybrid Helices
Hybrid DNA/RNA and triple helicies have been discussed in other places (such as PNA invasion), but are discussed here specifically because of their relevance (close relationship) to quadruplexes, and similar structures. Languages can be constructed for DNA/RNA hybrid triplexes too. The sample shape grammar (below) is crude in that triplexes tend to be composed of short sequences of bases, and may be far apart, and may be limited in the types that exist simultaneously, but the grammar is only to show possibilities.
Shape Grammar for DNA/RNA Hybrid Triple-Helices
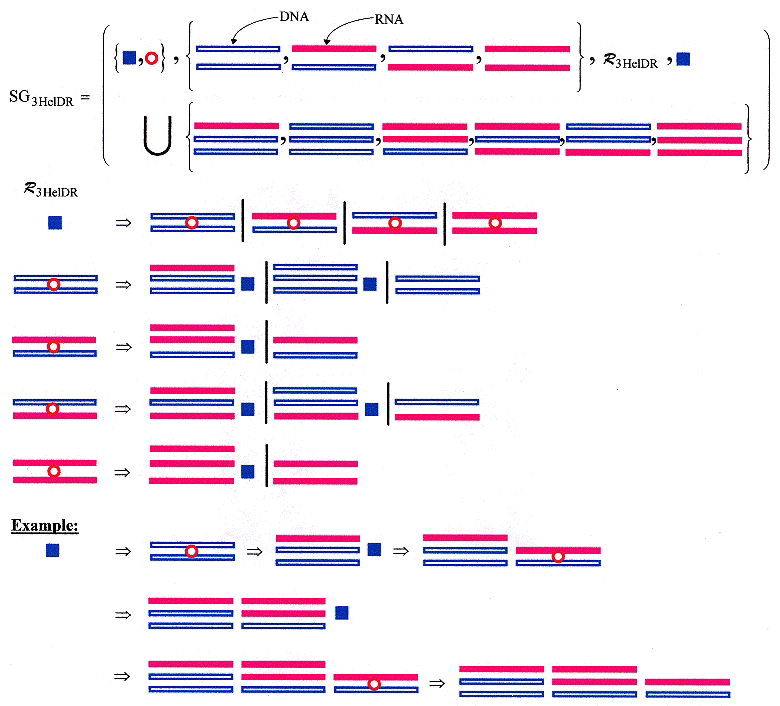
An alternative way to construct a grammar for triple-helix DNA is to use a Matrix or an Array Grammar" e, f, g, h, i, j.
A Matrix/Array grammar for DNA Triple-Helix
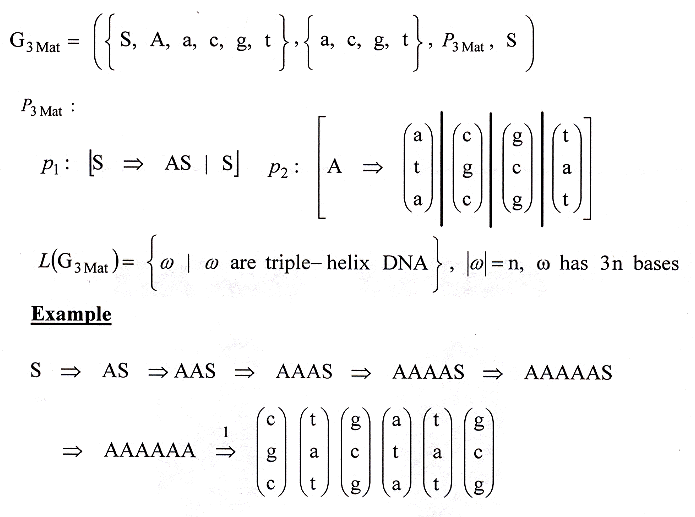
a "Triple-Helical Nucleic Acids", by V. N. Soyfer, V. N. Potaman, Springer-Verlag, New York, 1995
b "Binding of Two Different DNA Sequences by Conformational Switching", by E. Rubin, T. L. McKee, E. T, Kool, Journal of the American Chemical Society, Jan. 13 1993, 115, 1, 360 - 361
c "Sequence-specific recognition of a double helical RNA and RNA·DNA by triple helix formation", by H. Han, P. B. Dervan, Proceedings of the National Academy of Sciences USA, May 1 1993, 90, 9, 3806 - 3810
d "Stability and Properties of Double and Triple Helices: Dramatic Effects of RNA or DNA Backbone Composition", by R. W. Roberts, D. M. Crothers, Science, Nov. 27 1992, 258, 5087, 1463 - 1466
e "Abstract Families of Matrices and Picture Languages", by G. Siromoney, R. Siromoney, K. Krithivasan, Computer Grphics and Image Processing, 1972, 1, 284 - 307
f "Picture Languages with rray Rewriting Rules", by G. Siromoney, R. Siromoney, K. Krithivasan, Information and Control, 1973, 22, 447 - 470
g "On Equal Matrix Languages", by R. Siromoney, Information and Control, 1969, 14, 135 - 151
h "Channel Capacity of Equal Matrix Languages", by R. Siromoney, Information and Control, 1969, 14, 507 - 511
i "Hierarchical Structures and Complexities of Parallel Isometric Languages", by P. S. P. Wang, IEEE Transactions on Pattern Analysis and Machine Intelligence, Jan. 1983, 5, 1, 92 - 99
j "Array Grammars, Patterns, and Recognizers", P. S. P. Wang (Editor), World Scientific Publishers Co. Pte. Ltd., Singapore, 1989
"Targeted Cross-linking of Human β-Globin Gene in Living Cells Mediated by a Triple Helix Forming Oligonucleotide", by K. A. Shahid, A. Mujumdar, R. Alam, S.-T. Liu, J. Y. Kuan, X. Sui, B. Cuenoud, P. M. Glazar, P. S. Miller, M. M. Seidman, Biochemistry, 2006, 45, 1970 - 1978
This paper discusses Triple Helix Forming Oligonucleotides (TFOs).
"A gold nanoparticle based approach for screening triplex DNA binders", by Han, M. S., Lytton-Jean, A. K. R., Mirkin, C. A., Journal of the American Chemical Society, 19 April 2006, 128, 15, 4954 - 4955
Triplex forming oligonucleotides (TFOs) can be used as genetic-based disease pharmaceuticals. As drugs are developed, a fast and assured assay to test for TFO activity is desired. This paper describes a colorometric method to test for TFO activity using benzo[e]pyridoindole (BePI) or coralyne (CORA) to stabilize triplex Hoogsteen Py&midot;PuPy triple helices. Stabilized triplex allows gold nanoparticle aggregation which results in a red-shift, thus a colorometric detected change that is specific to triplexes.
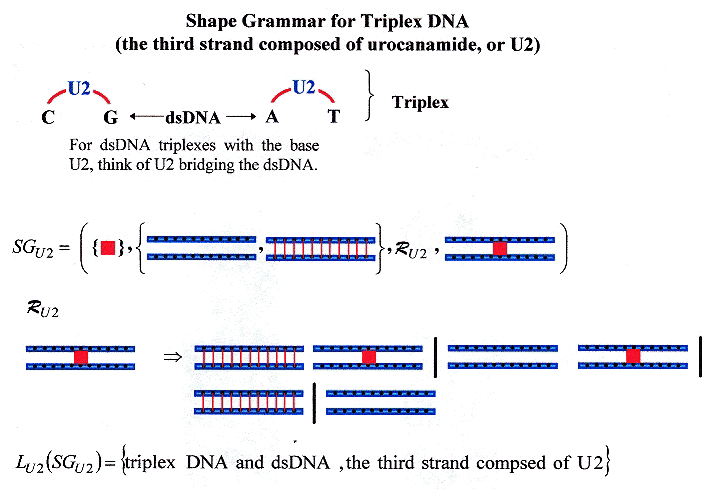
"Synthesis and DNA triplex formation of an oligonucleotide containing urocanamide", by M. G. M. Purwanto, K. Weisz, Tetrahedron Letters, June 5 2006, 47, 23, 3849 - 3852
A synthetic DNA triplex using urocanamide to bridge dsDNA thereby creating DNA Triplexes is reported.
Urocanamide DNA Triplexes
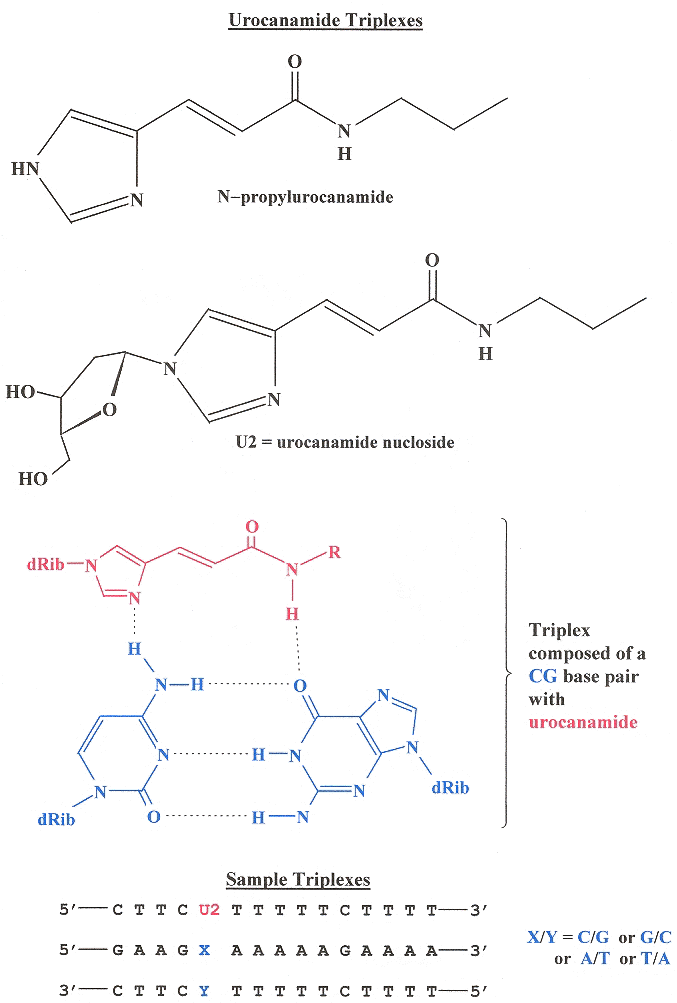
Shape Grammar for dsDNA mixed with Urocanamide DNA Triplexes
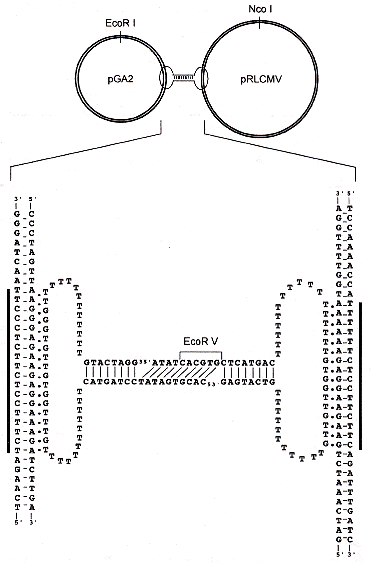
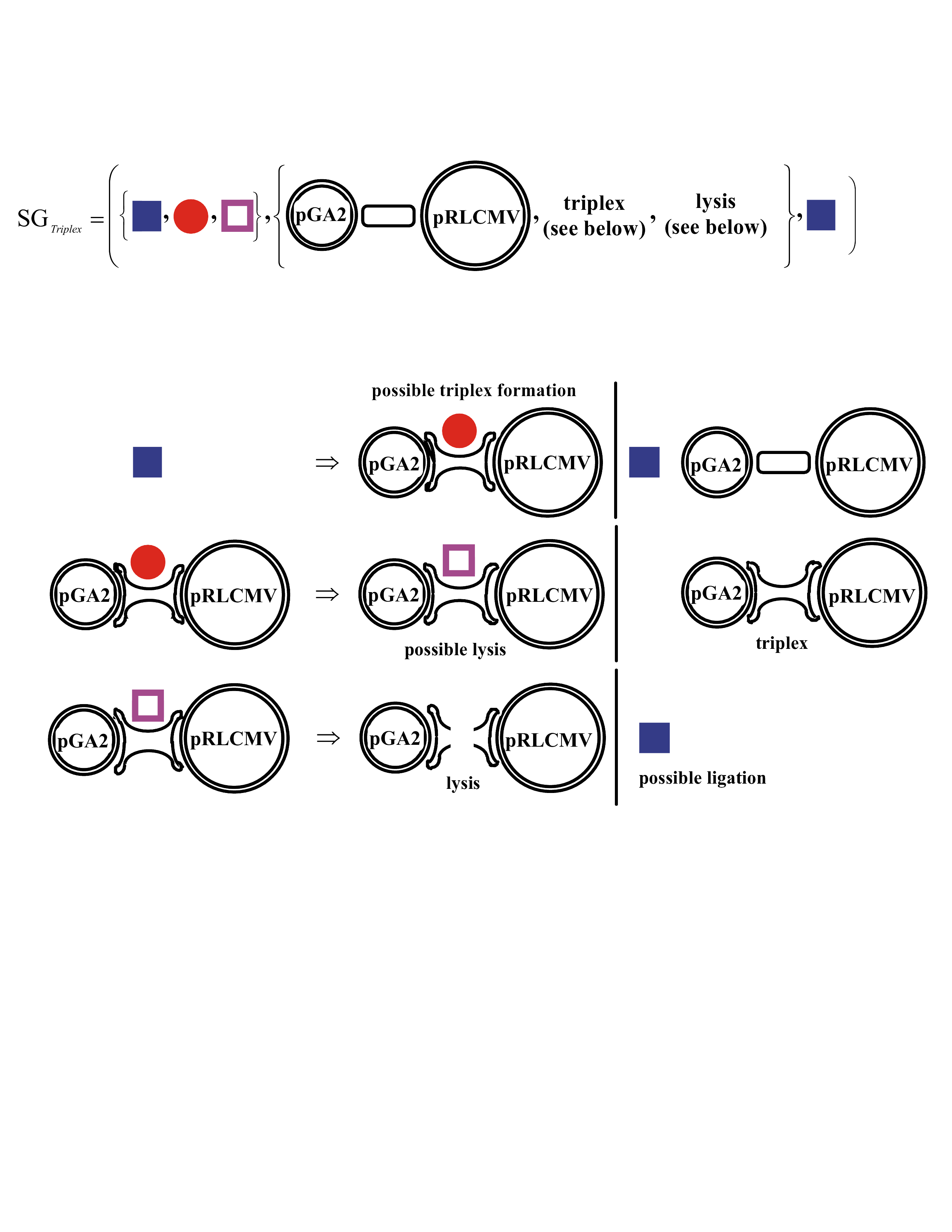
"A New Supramolecular Structure Made of Two Different Plasmids Linked by a Circular Oligonucleotide", by T. Roulon, E. Le Cam, C. Escudé, ChemBioChem, June 2006, 7, 6, 912 - 915
A new way to cleave triple-helices, found in the plasmid world is discussed. Two plasmids pGA2 and pRLCMV are linked by a circular oligonucleotide. The circular nucleotide binds as a triple-helix with each plasmid DNA.
For a review of relevant information concerning plasmids
A New Triple-Helix Cleavage Mechanism
A Shape Grammar for this New Triple-Helix Cleavage Mechanism
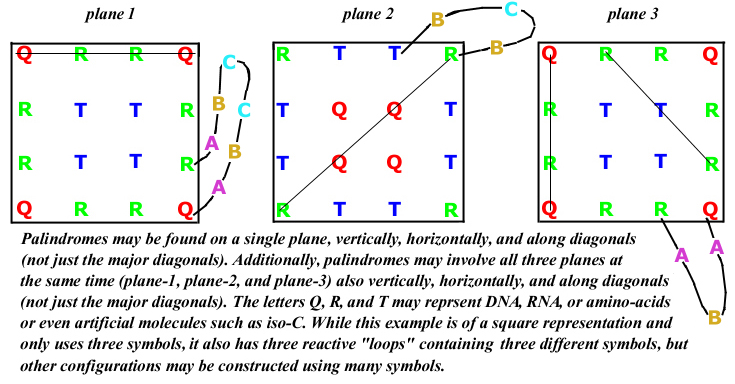
1-, 2-, and 3-Dimensional Palindromes
3-Dimensional Square Palindromes
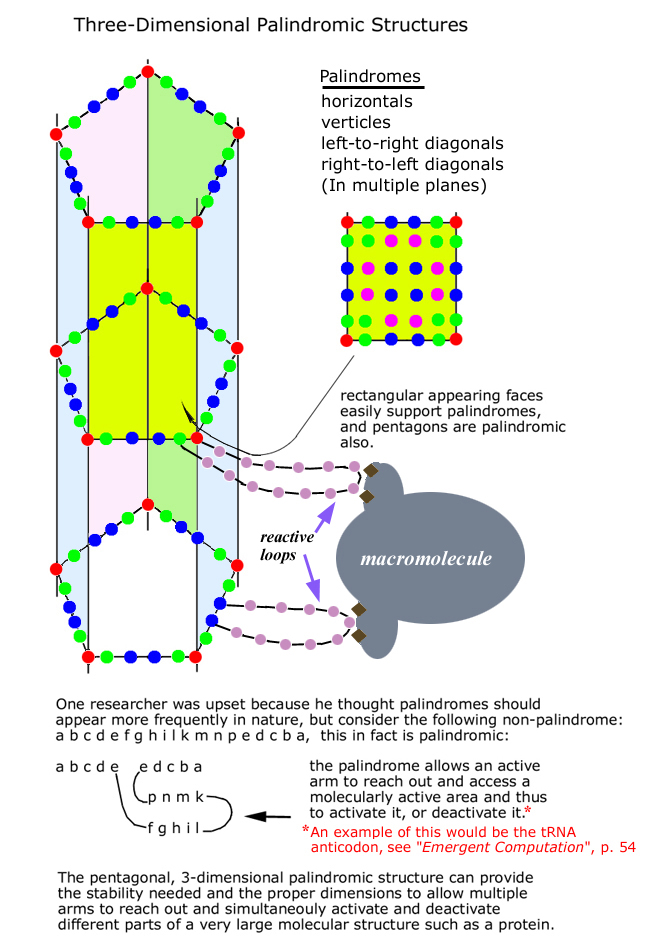
3-Dimensional Mixed Pentagonal and Rectangular Palindromes
© Matthew Simon, 2005 - 2017